Congenital tooth agenesis is a common anomaly in humans. We investigated the etiology of human tooth agenesis by exome analysis in Japanese patients, and found a previously undescribed heterozygous deletion (NM_002448.3(MSX1_v001):c.433_449del) in the first exon of the MSX1 gene. The deletion leads to a frameshift and generates a premature termination codon. The truncated form of MSX1, namely, p.(Trp145Leufs*24) lacks the homeodomain, which is crucial for transcription factor function.
Similar content being viewed by others
Congenital tooth agenesis is a common human anomaly classified into two subtypes based on the number of permanently missing teeth, excluding the third molars. Hypodontia is a condition defined as the absence of 1–5 permanent teeth (OMIM: #106600, #604625); oligodontia is another type of tooth agenesis in which six or more teeth are absent (OMIM: #167416). In most countries, ~5–10% of the total population is affected by congenital tooth agenesis, excluding the third molars1,2. Although oligodontia morbidity is more rarely observed, i.e., in 0.08–0.16% of the population, it shows high heritability3. A series of genetic studies have revealed the causative genes of human tooth agenesis, which include muscle segment homeobox1 (MSX1), paired box 9 (PAX9), ectodysplasin A (EDA), ectodysplasin A receptor (EDAR), and EDAR-associated death domain (EDARADD). In addition, several genes of the WNT/beta-Catenin signaling cascade, such as wingless-type MMTV integration site 10 A (WNT10A), low-density lipoprotein receptor-related protein 6 (LRP6), and axis inhibition protein 2 (AXIN2), are associated with human tooth malformation. These proteins play pivotal roles during early human development, including odontogenesis; thus, a mutation in one of the genes may cause both nonsyndromic and syndromic tooth agenesis4,5,6,7,8,9. MSX1 and PAX9 are homeoprotein transcription factors expressed in the dental mesenchyme with important roles in expression of mesenchymal bone morphogenetic protein 4 (BMP4), which promotes dental development10,11.
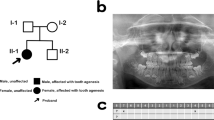
Here, we analyze Japanese patients with oligodontia diagnosed on the basis of clinical and radiographic examinations. Saliva samples were obtained from patients after they provided informed consent to participate in the study, which was approved by the Institutional Review Board of Aichi-Gakuin University, TOYOTA Memorial Hospital, and the Institute for Developmental Research, Aichi Developmental Disability Center. In the studied family, the proband (II-1) and her father (I-1) showed symptoms, i.e., loss of eight and six teeth, respectively (Fig. 1A–C). Nevertheless, we did not analyze the nucleotide sequence of the MSX1 gene of the father because of disagreement with informed consent. Orofacial cleft, craniofacial abnormalities, or other health problems, including those related to ectodermal organs, such as the hair, nails, and sweat glands, were not noted in any of the affected family members.
A The pedigree of the family with familial tooth agenesis. Squares and circles indicate male and female family members, respectively. Filled arrow indicates the proband. Black symbols indicate individuals who were clinically diagnosed with isolated tooth agenesis. B Panoramic tomogram of the proband, patient II-1. C Missing tooth pattern of the proband (II-1).
Genomic DNA was extracted from saliva using Oragene DNA (OG-500; DNA Genotek, Ontario, Canada) according to the manufacturer’s instructions. Briefly, each saliva sample was mixed with prepIT L2P (PT-2LP; DNA Genotek), incubated on ice, and centrifuged for 5 min at 15,000 × g. The supernatant was mixed with EtOH to precipitate DNA. After centrifugation, the DNA pellet was dissolved in elution buffer. Mutational analysis by whole-exome sequencing was subsequently performed according to our previously reported method12.
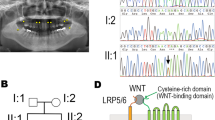
Whole-exome sequencing identified a novel, heterozygous, 17-base pair (TGGATGCAGAGCCCCCG) deletion in the first exon of the MSX1 gene (NM_002448.3(MSX1_v001):c.433_449del; c.1 is the A of the ATG translation initiation codon of the MSX1 mRNA; NM_002448.3 in the GenBank database; Fig. 2A). This mutation was not present in the following online databases: dbSNP, 1000 Genomes, and NHLBI exome variant project (http://evs.gs.washington.edu/EVS/). The deletion results in an amino acid substitution of the 145th tryptophan (NP_002439.2) to leucine, and the stop codon following an unrelated peptide sequence consists of 23 amino acid residues (NM_002448.3(MSX1_i001):p.(Trp145Leufs*24); Fig. 2B). Whole-exome sequencing data for the DNA samples did not involve other known causative genes for nonsyndromic tooth agenesis, e.g., PAX9, WNT10A, LRP6, EDA1, PITX2, AXIN2, EDA1, EDAR, and EDARADD.
A Sequencing results for the MSX1 gene in the control (upper panel). Heterologous peaks of the nucleotide sequence for tooth agenesis were detected in the patient’s MSX1 gene (lower panel). B Schematic diagram of wild-type MSX1 and the mutant; p.(Trp145Leufs*24). The MSX1 mutant is a C-terminus-truncated form lacking the homeodomain/MH4 and the PIAS-binding domain. PBD PIAS-binding domain (amino acids 265–297). C Nuclear localization of wild-type and truncated MSX1 in HEK293 cells. FLAG-tagged wild-type MSX1 immunolocalized to the nucleus of transfected HEK293 cells, but immunoreactivity of the mutant MSX1 was diffusely detected in the cytoplasm. MSX1 (FLAG, red); nuclei [4′,6-diamidino-2-phenylindole (DAPI), blue]. Bar = 10 µm.
The 17-bp deletion generates an MSX1 protein with a C-terminus lacking the homeodomain. It has previously been demonstrated that the homeodomain (amino acids 175–229) plays a pivotal role in molecular interactions with DNA13 and other transcription factors related to tooth development, such as PAX9, TATA-binding protein, and DLX family members14. The homeodomain is also associated with nuclear transport15, which is crucial for the biological function of transcription factors. We expressed mutant MSX1 in HEK293 cells to confirm the nuclear transport defect. Although immunoreactivity of wild-type MSX1 was detected in nuclei, as previously reported16, truncated MSX1 was mainly found in the cytoplasm (Fig. 2C).
In addition to its nonsyndromic form, haploinsufficiency of MSX1 causes the syndromic form of tooth agenesis that includes cleft lip and/or palate17,18. To date, 50 MSX1 gene mutations, including four truncated variants, have been identified in patients with syndromic and nonsyndromic tooth agenesis. Although amino acid substitutions in patients with tooth agenesis cluster in the homeodomain (70%), no variants associated with cleft lip and/or palate without tooth malformation have been identified in it19. Previous biochemical analyses have shown that some single-amino acid substitutions in the homeodomain of MSX1 can affect the transcriptional suppression activity of the MyoD promoter, which is one of the targets of MSX120. Because MSX1 strongly suppresses target gene expression and represses cell differentiation both in vitro and in vivo21, it sustains cellular proliferation in the tooth germ during odontogenesis for robust tooth development in terms of size, number, and shape.
In summary, previous reports and the current results indicate that the p.(Trp145Leufs*24) mutation impairs the molecular function of MSX1; thus, the identified nucleotide deletion is the cause of the tooth agenesis in the studied family. Because of the pathogenic relationship between particular MSX1 mutations and facial clefts, we believe that clarifying MSX1 gene variations will help to improve the precision of genetic counseling to patients with odontogenic malformations and/or facial clefts.
HGV Database
The relevant data from this Data Report are hosted at the Human Genome Variation Database at https://doi.org/10.6084/m9.figshare.hgv.3039
References
Polder, B. J. et al. A meta‐analysis of the prevalence of dental agenesis of permanent teeth. Community Dent. Oral Epidemiol. 32, 217–226 (2004).
Fekonja, A. Hypodontia in orthodontically treated children. Eur. J. Orthod. 27, 457–460 (2005).
Rølling, S. & Poulsen, S. Oligodontia in Danish schoolchildren. Acta Odontol. Scand. 59, 111–112 (2001).
Vastardis, H. et al. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat. Genet. 13, 417–421 (1996).
Stockton, D. W. et al. Mutation of PAX9 is associated with oligodontia. Nat. Genet. 24, 18–19 (2000).
Lammi, L. et al. Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am. J. Hum. Genet. 74, 1043–1050 (2004).
Machida, J. et al. WNT10A variants isolated from Japanese patients with congenital tooth agenesis. Hum. Genome Var. 4, 1–4 (2017).
Massink, M. P. et al. Loss-of-function mutations in the WNT co-receptor LRP6 cause autosomal-dominant oligodontia. Am. J. Hum. Genet. 97, 621–626 (2015).
Song, S. et al. EDA gene mutations underlie non-syndromic oligodontia. J. Dent. Res. 88, 126–131 (2009).
Satokata, I. & Maas, R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat. Genet. 6, 348–356 (1994).
Peters, H. et al. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 12, 2735–2747 (1998).
Tatematsu, T. et al. An aberrant splice acceptor site due to a novel intronic nucleotide substitution in MSX1 gene is the cause of congenital tooth agenesis in a Japanese family. PLoS ONE 10, e0128227 (2015).
Hovde, S. et al. Crystal structure of the Msx-1 homeodomain/DNA complex. Biochemistry 40, 12013–12021 (2001).
Hu, G. et al. Haploinsufficiency of MSX1: a mechanism for selective tooth agenesis. Mol. Cell. Biol. 18, 6044–6051 (1998).
Wang, Y. et al. Msx1 mutations: how do they cause tooth agenesis? J. Dent. Res. 90, 311–316 (2011).
Lee, H. et al. PIAS1 confers DNA-binding specificity on the Msx1 homeoprotein. Genes Dev. 20, 784–794 (2006).
Jezewski, P. A. et al. Complete sequencing shows a role for MSX1 in non-syndromic cleft lip and palate. J. Med. Genet. 40, 399–407 (2003).
Finnerty, J. R. et al. Domain duplication, divergence, and loss events in vertebrate Msx paralogs reveal phylogenomically informed disease markers. BMC Evol. Biol. https://doi.org/10.1186/1471-2148-9-18 (2009).
Bonczek, O. et al. Next generation sequencing reveals a novel nonsense mutation in MSX1 gene related to oligodontia. PLoS ONE 13, e0202989 (2018).
Kamamoto, M. et al. Clinical and functional data implicate the Arg (151) Ser variant of MSX1 in familial hypodontia. Eur. J. Hum. Genet. 19, 844–850 (2011).
Wang, J. et al. The Msx1 homeoprotein recruits polycomb to the nuclear periphery during development. Dev. Cell 21, 575–588 (2011).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Adachi, J., Aoki, Y., Tatematsu, T. et al. Novel MSX1 frameshift mutation in a Japanese family with nonsyndromic oligodontia. Hum Genome Var 8, 29 (2021). https://doi.org/10.1038/s41439-021-00161-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41439-021-00161-x
This article is cited by
-
Novel frameshift variant of WNT10A in a Japanese patient with hypodontia
Human Genome Variation (2024)
-
Novel WNT10A variant in a Japanese case of nonsyndromic oligodontia
Human Genome Variation (2023)