Abstract
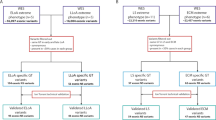
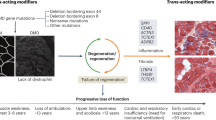
The major determinant of disease severity in Duchenne muscular dystrophy (DMD) or milder Becker muscular dystrophy (BMD) is whether the dystrophin gene (DMD) mutation truncates the mRNA reading frame or allows expression of a partially functional protein. However, even in the complete absence of dystrophin, variability in disease severity is observed, and candidate gene studies have implicated several genes as modifiers. Here we present the largest genome-wide search to date for loci influencing severity in N = 419 DMD patients. Availability of subjects for such studies is quite limited, leading to modest sample sizes, which present a challenge for GWAS design. We have therefore taken special steps to minimize heterogeneity within our dataset at the DMD locus itself, taking a novel approach to mutation classification to effectively exclude the possibility of residual dystrophin expression, and utilized statistical methods that are well adapted to smaller sample sizes, including the use of a novel linear regression-like residual for time to ambulatory loss and the application of evidential statistics for the GWAS approach. Finally, we applied an unbiased in silico pipeline, utilizing functional genomic datasets to explore the potential impact of the best supported SNPs. In all, we obtained eight SNPs (out of 1,385,356 total) with posterior probability of trait-marker association (PPLD) ≥ 0.4, representing six distinct loci. Our analysis prioritized likely non-coding SNP regulatory effects on six genes (ETAA1, PARD6G, GALNTL6, MAN1A1, ADAMTS19, and NCALD), each with plausibility as a DMD modifier. These results support both recurrent and potentially new pathways for intervention in the dystrophinopathies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Flanigan KM, Dunn DM, von Niederhausern A, Soltanzadeh P, Howard MT, Sampson JB, et al. Nonsense mutation-associated Becker muscular dystrophy: interplay between exon definition and splicing regulatory elements within the DMD gene. Hum Mutat. 2011;32:299–308.
Bello L, Flanigan KM, Weiss RB, Spitali P, Aartsma-Rus A, Muntoni F, et al. Association study of exon variants in the NF-kappaB and TGFbeta pathways identifies CD40 as a modifier of duchenne muscular dystrophy. Am J Hum Genet. 2016;99:1163–71.
Bello L, Pegoraro E. The “Usual Suspects”: genes for inflammation, fibrosis, regeneration, and muscle strength modify duchenne muscular dystrophy. J Clin Med. 2019;8:649.
Weiss RB, Vieland VJ, Dunn DM, Kaminoh Y, Flanigan KM, United Dystrophinopathy P. Long-range genomic regulators of THBS1 and LTBP4 modify disease severity in duchenne muscular dystrophy. Ann Neurol. 2018;84:234–45.
Heydemann A, Ceco E, Lim JE, Hadhazy M, Ryder P, Moran JL, et al. Latent TGF-beta-binding protein 4 modifies muscular dystrophy in mice. J Clin Invest. 2009;119:3703–12.
Flanigan KM, Ceco E, Lamar KM, Kaminoh Y, Dunn DM, Mendell JR, et al. LTBP4 genotype predicts age of ambulatory loss in Duchenne muscular dystrophy. Ann Neurol. 2013;73:481–8.
Pegoraro E, Hoffman EP, Piva L, Gavassini BF, Cagnin S, Ermani M, et al. SPP1 genotype is a determinant of disease severity in Duchenne muscular dystrophy. Neurology. 2011;76:219–26.
Quattrocelli M, Capote J, Ohiri JC, Warner JL, Vo AH, Earley JU, et al. Genetic modifiers of muscular dystrophy act on sarcolemmal resealing and recovery from injury. PLoS Genet. 2017;13:e1007070.
Vieira NM, Elvers I, Alexander MS, Moreira YB, Eran A, Gomes JP, et al. Jagged 1 rescues the duchenne muscular dystrophy phenotype. Cell. 2015;163:1204–13.
Crawford SE, Stellmach V, Murphy-Ullrich JE, Ribeiro SM, Lawler J, Hynes RO, et al. Thrombospondin-1 is a major activator of TGF-beta1 in vivo. Cell. 1998;93:1159–70.
Spitali P, Zaharieva I, Bohringer S, Hiller M, Chaouch A, Roos A, et al. TCTEX1D1 is a genetic modifier of disease progression in Duchenne muscular dystrophy. Eur J Hum Genet. 2020;28:815–25.
Vieland VJ, Seok SC. The PPLD has advantages over conventional regression methods in application to moderately sized genome-wide association studies. PLoS One. 2021;16:e0257164.
Flanigan KM, Dunn DM, von Niederhausern A, Soltanzadeh P, Gappmaier E, Howard MT, et al. Mutational spectrum of DMD mutations in dystrophinopathy patients: application of modern diagnostic techniques to a large cohort. Hum Mutat. 2009;30:1657–66.
Flanigan KM, von Niederhausern A, Dunn DM, Alder J, Mendell JR, Weiss RB. Rapid direct sequence analysis of the dystrophin gene. Am J Hum Genet. 2003;72:931–9.
Waldrop MA, Gumienny F, El Husayni S, Frank DE, Weiss RB, Flanigan KM. Low-level dystrophin expression attenuating the dystrophinopathy phenotype. Neuromuscul Disord. 2018;28:116–21.
Flanigan KM, Dunn DM, von Niederhausern A, Howard MT, Mendell J, Connolly A, et al. DMD Trp3X nonsense mutation associated with a founder effect in North American families with mild Becker muscular dystrophy. Neuromuscul Disord. 2009;19:743–8.
Muntoni F, Gobbi P, Sewry C, Sherratt T, Taylor J, Sandhu SK, et al. Deletions in the 5’ region of dystrophin and resulting phenotypes. J Med Genet. 1994;31:843–7.
Wang RT, Barthelemy F, Martin AS, Douine ED, Eskin A, Lucas A, et al. DMD genotype correlations from the Duchenne Registry: endogenous exon skipping is a factor in prolonged ambulation for individuals with a defined mutation subtype. Hum Mutat. 2018;39:1193–202.
Shiga N, Takeshima Y, Sakamoto H, Inoue K, Yokota Y, Yokoyama M, et al. Disruption of the splicing enhancer sequence within exon 27 of the dystrophin gene by a nonsense mutation induces partial skipping of the exon and is responsible for Becker muscular dystrophy. J Clin Investig. 1997;100:2204–10.
Disset A, Bourgeois CF, Benmalek N, Claustres M, Stevenin J, Tuffery-Giraud S. An exon skipping-associated nonsense mutation in the dystrophin gene uncovers a complex interplay between multiple antagonistic splicing elements. Hum Mol Genet. 2006;15:999–1013.
Vieland VJ, Huang Y, Seok S-C, Burian J, Catalyurek U, O’Connell J, et al. KELVIN: a software package for rigorous measurement of statistical evidence in human genetics. Hum Hered. 2011;72:276–88.
Vieland VJ, Seok S-C, Stewart WCL. A new linear regression-like residual for survival analysis, with application to genome wide association studies of time-to-event data. PLoS One. 2020;15:e0232300.
Miosge LA, Sontani Y, Chuah A, Horikawa K, Russell TA, Mei Y, et al. Systems-guided forward genetic screen reveals a critical role of the replication stress response protein ETAA1 in T cell clonal expansion. Proc Natl Acad Sci USA. 2017;114:E5216–25.
Dumont NA, Wang YX, von Maltzahn J, Pasut A, Bentzinger CF, Brun CE, et al. Dystrophin expression in muscle stem cells regulates their polarity and asymmetric division. Nat Med. 2015;21:1455–63.
Yamashita K, Suzuki A, Satoh Y, Ide M, Amano Y, Masuda-Hirata M, et al. The 8th and 9th tandem spectrin-like repeats of utrophin cooperatively form a functional unit to interact with polarity-regulating kinase PAR-1b. Biochem Biophys Res Commun. 2010;391:812–7.
Feige P, Brun CE, Ritso M, Rudnicki MA. Orienting muscle stem cells for regeneration in homeostasis, aging, and disease. Cell Stem Cell. 2018;23:653–64.
Bankole LC, Feasson L, Ponsot E, Kadi F. Fibre type-specific satellite cell content in two models of muscle disease. Histopathology. 2013;63:826–32.
Chang NC, Chevalier FP, Rudnicki MA. Satellite cells in muscular dystrophy—lost in polarity. Trends Mol Med. 2016;22:479–96.
Vierstra J, Lazar J, Sandstrom R, Halow J, Lee K, Bates D, et al. Global reference mapping of human transcription factor footprints. Nature. 2020;583:729–36.
Rodriguez-Martinez JA, Reinke AW, Bhimsaria D, Keating AE, Ansari AZ. Combinatorial bZIP dimers display complex DNA-binding specificity landscapes. Elife. 2017;6:e19272.
Nasser J, Bergman DT, Fulco CP, Guckelberger P, Doughty BR, Patwardhan TA, et al. Genome-wide enhancer maps link risk variants to disease genes. Nature. 2021;593:238–43.
De Micheli AJ, Laurilliard EJ, Heinke CL, Ravichandran H, Fraczek P, Soueid-Baumgarten S, et al. Single-cell analysis of the muscle stem cell hierarchy identifies heterotypic communication signals involved in skeletal muscle regeneration. Cell Rep. 2020;30:3583–95.e3585.
Chemello F, Wang Z, Li H, McAnally JR, Liu N, Bassel-Duby R, et al. Degenerative and regenerative pathways underlying Duchenne muscular dystrophy revealed by single-nucleus RNA sequencing. Proc Natl Acad Sci USA. 2020;117:29691–701.
Sacco A, Mourkioti F, Tran R, Choi J, Llewellyn M, Kraft P, et al. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell. 2010;143:1059–71.
Latella L, Dall’Agnese A, Boscolo FS, Nardoni C, Cosentino M, Lahm A, et al. DNA damage signaling mediates the functional antagonism between replicative senescence and terminal muscle differentiation. Genes Dev. 2017;31:648–59.
Wang YX, Feige P, Brun CE, Hekmatnejad B, Dumont NA, Renaud JM, et al. EGFR-Aurka signaling rescues polarity and regeneration defects in dystrophin-deficient muscle stem cells by increasing asymmetric divisions. Cell Stem Cell. 2019;24:419–32.e416.
Dormoy V, Tormanen K, Sutterlin C. Par6gamma is at the mother centriole and controls centrosomal protein composition through a Par6alpha-dependent pathway. J Cell Sci. 2013;126:860–70.
Fahed AC, Wang M, Homburger JR, Patel AP, Bick AG, Neben CL, et al. Polygenic background modifies penetrance of monogenic variants for tier 1 genomic conditions. Nat Commun. 2020;11:3635.
Acknowledgements
The data used for the analyses described in this manuscript were obtained from the GTEx Portal using the GTEx Analysis Release V8 (dbGaP Accession phs000424.v8.p2). The authors wish to acknowledge the many members of the United Dystrophinopathy Project consortium who participated in the enrollment of subjects in the original historical UDP database.
Funding
This work was supported by the National Institutes of Health (NINDS NS085238) to KMF, RBW, and VJV. The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS.
Author information
Authors and Affiliations
Contributions
RBW, VJV, and KMF conceived and designed the study. RBW, VJV, DMD, MW, RA, LA, MM-C, KMF, JB, SCS, and the UDP Investigators acquired and/or analyzed the data. RBW, VJV, PTM, and KMF drafted the manuscript, which was reviewed and revised by all of the named authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The research involving human subjects has been been performed in accordance with the Declaration of Helsinki and was approved by the Nationwide Children’s Hospital Institutional Review Board (IRB) under approval 0502HSE046. Informed consent was obtained from all subjects.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Flanigan, K.M., Waldrop, M.A., Martin, P.T. et al. A genome-wide association analysis of loss of ambulation in dystrophinopathy patients suggests multiple candidate modifiers of disease severity. Eur J Hum Genet 31, 663–673 (2023). https://doi.org/10.1038/s41431-023-01329-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41431-023-01329-5
This article is cited by
-
2023 in the European Journal of Human Genetics
European Journal of Human Genetics (2024)
-
Is it time for genetic modifiers to predict prognosis in Duchenne muscular dystrophy?
Nature Reviews Neurology (2023)
-
The complex genomics of single gene disorders
European Journal of Human Genetics (2023)