Abstract
Macrophages play a critical role in the immune homeostasis and host defense against invading pathogens. However, uncontrolled activation of inflammatory macrophages leads to tissue injury and even fuels autoimmunity. Hence the molecular mechanisms underlying macrophage activation need to be further elucidated. The effects of epigenetic modifications on the function of immune cells draw increasing attention. Here, we demonstrated that lysine-specific demethylase 5B (KDM5B), a classical transcriptional repressor in stem cell development and cancer, was required for the full activation of NF-κB signaling cascade and pro-inflammatory cytokine production in macrophages. KDM5B deficiency or inhibitor treatment protected mice from immunologic injury in both collagen-induced arthritis (CIA) model and endotoxin shock model. Genome-wide analysis of KDM5B-binding peaks identified that KDM5B was selectively recruited to the promoter of Nfkbia, the gene encoding IκBα, in activated macrophages. KDM5B mediated the H3K4me3 modification erasing and decreased chromatin accessibility of Nfkbia gene locus, coordinating the elaborate suppression of IκBα expression and the enhanced NF-κB-mediated macrophage activation. Our finding identifies the indispensable role of KDM5B in macrophage-mediated inflammatory responses and provides a candidate therapeutic target for autoimmune and inflammatory disorders.
Similar content being viewed by others
Introduction
Macrophage is the master regulator of the induction and resolution of host inflammation in infection and autoimmunity [1, 2]. Generally, pattern recognition receptors (PRRs) on macrophages specifically recognize and bind the pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DMAPs) for launching the downstream signaling cascades, including nuclear factor kappa-B (NF-κB) pathway, mitogen-activated protein kinase (MAPK) pathway, and etc [3]. Upon activated, macrophages robustly produce pro-inflammatory cytokine (e.g., IL-1β, IL-6, IL-12, and TNF-α) for the initiation of innate immune responses [4]. Otherwise, the co-stimulatory signals and pro-inflammatory mediators produced by macrophages set up the immune environment for effective mobilization of adaptive immune responses, such as the potent T helper type 1 (Th1)-Th17 response [5]. In this way, resident and infiltrating macrophages in the inflamed tissues, together with T cells, B cells and other inflammatory cells, disrupt the immune homeostasis and result in autoimmune disorders. Rheumatoid arthritis (RA) is a chronic autoimmune disease involving joint and surrounding tissues with uncontrolled inflammatory responses. Synovial hyperplasia and excessive immune cell accumulation cause the undesirable tissue remodeling and reduced function of involved joints [6]. RA patients have increased number of active inflammatory macrophages, which were widely known as the central players in the pathogenesis of RA [7, 8]. Therefore, increasing studies focus on the therapeutic intervention for preventing the pathological effects of inflammatory macrophages.
Histone modification, mainly including histone methylation, acetylation, phosphorylation, and ubiquitination, is well accepted as the important regulator of gene expression and cell destiny, whose dysfunction leads to pathological conditions and even severe diseases. Increasing researchers found that various histone modifications participated in the regulation of macrophage development and function, which might be the promising therapeutic target for macrophage-associated diseases [9]. For example, in the late phase of macrophage inflammatory polarization, cellular metabolism reprogramming in macrophages induced histone lactylation and the enhanced expression of homeostatic genes involving wound healing, such as arginase 1 (Arg1) [10]. Recent work found that mitochondrial glucose oxidation and ATP citrate lyase (ACLY)-dependent acetyl-CoA generation was the rate-limiting step in the histone 3 lysine 9 (H3K9) acetylation-driven macrophage activation and inflammatory gene expression in vivo [11]. However, more biological functions of specific histone modifications, along with the “writer” or “erasers” for these modifications, in macrophages and autoimmune diseases remain to be further explored.
Lysine-specific demethylase 5B (KDM5B), also named as Jumonji AT-rich interactive domain 1B (JARID1B) or PLU-1, belongs to the jumonji/ARAD (Jmjd) domain-containing family of histone demethylases. KDM5B is highly expressed in a large variety of cancers, especially breast cancer, colorectal cancer, gastric carcinoma and etc. [12]. KDM5B was reported to function as the transcription repressor through removing promoter-associated histone 3 lysine 4 trimethylation (H3K4me3) in kinds of cells. KDM5B regulated the expression of CC chemokine ligand 14 (CCL14) in HT-29 cells and cyclin dependent kinase inhibitor 1A (CDKN1A) in MCF-7 cells through removing the H3K4me3 modification at the gene promoters [13,14,15]. In addition, deletion of KDM5B in cancer cells was required for melanoma-induced adaptive immune responses and enhanced responses to immune checkpoint blockade, which was not dependent on demethylase activity but on the interaction with SET domain bifurcated histone lysine methyltransferase 1 (SETDB1) for silencing retroelements-triggered type I interferon (IFN-I) responses [16]. Hence KDM5B regulated gene expression in both demethylase-dependent or -independent manner, according to different settings. However, the specific role of KDM5B in immune cells, especially in macrophages, remains largely unknown.
In this study, we found the significant decreased expression of KDM5B in synovial macrophages from RA patients. KDM5B deficiency ameliorated the pathological injury and immune disorder in collagen-induced arthritis (CIA) model and endotoxin shock model. KDM5B was essential for the production of pro-inflammatory cytokines including IL-1β, IL-6 and TNF-α in macrophages through maintaining the NF-κB signaling cascade activation. LPS stimulation induced the specific recruitment of KDM5B to the promoter of Nfkbia and mediated its transcriptional repression, while KDM5B deficiency led to the increased IκBα protein level that blocked NF-κB transcriptional activity. Our study reveals that KDM5B functions as an indispensable epigenetic regulator in macrophage activation and inflammatory disorders.
Results
The expression of histone demethylase KDM5B is extensively decreased in multiple autoimmune diseases
To identify the regulatory protein involving in the inflammatory response and autoimmune diseases, we searched the Gene Expression Omnibus (GEO) database and further analyzed the differentially expressed genes (DEGs) in synovial macrophages from health control and RA patients in GSE97779 [17] (Supplementary Fig. 1A). In synovial macrophages from RA group, the expression of several histone demethylase family members showed a dramatic difference from those in health control groups (Supplementary Fig. 1B). Among them, KDM5B that was significantly down-regulated in RA patients drew our attention (Supplementary Fig. 1C). We next analyzed the expression of KDM5B in other autoimmune diseases including multiple sclerosis (MS), inflammatory bowel diseases (IBD) and cryopyrin-associated periodic syndromes (CAPS), all of which involving the aberrant activation of macrophages and macrophage-derived excessive pro-inflammatory responses. KDM5B expression was also significantly reduced in the PBMCs from MS patients, compared with that from health control (Supplementary Fig. 1D) [18]. Additionally, KDM5B was down-regulated in the PBMCs and inflamed colonic mucosa samples from ulcerative colitis (UC) patients (Supplementary Fig. 1E, F) [19, 20]. We also found the significant reduction of KDM5B expression in whole peripheral blood from CAPS patients (Supplementary Fig. 1G) [21]. To get a more detail understanding of KDM5B expression in RA patients, we collected the PBMCs from health control, stable RA (SRA) patients and active RA (ARA) patients, who were qualified for the diagnostic criteria set by American College of Rheumatology [22]. Compared with that in HC, KDM5B expression was significantly repressed in ARA patients on both mRNA and protein levels (Supplementary Fig. 1H–J). The above data imply that KDM5B has potential biological functions in the pathogenesis of these autoimmune diseases.
Next, we investigated the expression change of KDM5B in macrophages subjected to different stimuli. The data from GSE19315 showed that KDM5B expression was decreased in macrophage-like THP-1 cells treated with LPS, the ligand of Toll-like receptor 4 (TLR4) (Supplementary Fig. 2A) [23]. As presented in data from GSE5009, KDM5B expression was suppressed in IFN-γ- and LPS-treated macrophages but not in IL-4-treated murine macrophages (Supplementary Fig. 2B) [24]. Our experimental data also showed that the mRNA expression level of KDM5B was time-dependently down-regulated in mouse BMDMs stimulated with LPS, dsRNA analog poly(I:C) or gram-positive bacteria component Pam3CSK4 (Supplementary Fig. 2C–E). Nevertheless, Kdm5b mRNA expression in mouse BMDMs was hardly influenced by IL-4 treatment (Supplementary Fig. 2F). KDM5B protein expression was also suppressed in mouse BMDMs stimulated with LPS, poly(I:C) or Pam3CSK4 but not IL-4 (Supplementary Fig. 2G–J). For exploring the mechanisms underlying the decreased expression of KDM5B in LPS-activated macrophages, we analyzed the histone modification and RNA Pol II activity at the KDM5B promoter in BMDMs and found that LPS stimulation as long as 1 h (a short time) could inhibit the H3K27ac and RNA Pol II binding as well as S2P activity within the KDM5B gene locus, which were the markers of gene transcriptional activation (Supplementary Fig. 2K) [25, 26]. In addition, we analyzed the newly synthesis Kdm5b mRNA by GRO-seq to access the nascent RNA, and found that LPS stimulation resulted in the decreased Kdm5b nascent RNA production (Supplementary Fig. 2K) [26]. These results suggest that KDM5B expression is suppressed by a variety of inflammatory stimuli owing to the transcriptional inhibition, providing the potential explanation for the low expression in PBMCs or local inflamed tissues in patients with autoimmune diseases.
Loss of KDM5B ameliorates the immunologic injury in experimental arthritis model
To exploring the in vivo function of KDM5B, we generated the KDM5B knockout (KO) mice via CRISPR/Cas9 technology (Supplementary Fig. 3A, B). Compared with WT littermates, KDM5B-KO mice had normal physical development and comparable body weight (Supplementary Fig. 3C, D), despite of the low fertility rate which was consistent with the previous studies [27, 28]. We further observed the development of T lymphocytes in the thymus and peripheral immune organs from KDM5B-KO mice by FACS. KDM5B deficiency did not influence the percentages of double-negative (DN), double-positive (DP) and single-positive CD4+ and CD8+ T cell populations in thymus (Supplementary Fig. 3E). Meanwhile, KDM5B deficiency had no effect on the populations of CD19+ B cells and CD3+ T cells, as well as CD4+ and CD8+ T cells in spleen and lymphonodus (Supplementary Fig. 3F, G). Taken together, KDM5B-deficient mice develop the functional immune system and normal adaptive immune response, at least at the stable state.
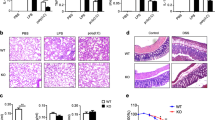
CIA model was taken to access the biological function of KDM5B in vivo, which was a widely-accepted murine inflammatory model with pathological characteristic similar to human RA patients [29]. Intriguingly, KDM5B-KO mice showed the relieved development of inflammatory arthritis, with delayed onset of disease, lower clinical scores, less ankle joint diameter and milder joint swelling at the front and hind paws, compared with WT mice (Fig. 1A–C; Supplementary Fig. 3H). Histological analysis of knee joints by H&E staining showed that the dramatic ameliorated disease severity with the reduced joint capsule swelling, attenuated synovial hyperplasia and less inflammatory cell infiltration in KDM5B-KO mice (Fig. 1D). In addition, safranin O staining turned out that cartilage and new bone destruction was prevented by KDM5B deficiency (Fig. 1E). The level of pro-inflammatory cytokine in sera including IL-1β, IL-6, IL-12p70 and TNF-α from KO mice was significantly lower than that from WT mice (Fig. 1F). These pro-inflammatory mediators derived from innate immune cells such as macrophages and dendritic cells (DCs) could result in the proliferation and differentiation of CD4+ T cells, contributing to the disrupted immune homeostasis and joint inflammation in the pathogenesis of RA [5, 30]. Therefore, we analyzed the pathogenic and regulatory T cell population in CIA mice and found that KDM5B-KO mice had the much lower percentages of pro-inflammatory Th1 and Th17 cells (Fig. 1G), which might facilitate bone erosion and destruction induced by collagen immunization. However, there was no difference in the population of CD25+FoxP3+ regulatory T (Treg) cells and CD4+CXCR5+PD-1+ follicular helper T (Tfh) cells in the spleen between WT and KO mice (Supplementary Fig. 3I, J). These data indicate that KDM5B deficiency protects mice from the pathologic injury and joint inflammation with less pro-inflammatory cytokine production in arthritis model.
A Clinical scores of littermate control WT and KDM5B-deficient (KO) mice after the 2nd immunization of CIA model (n = 6 mice per group). B Diameter changes in ankle joints of WT and KO mice on day 14 and day 28 after the 2nd immunization (n = 6 mice per group). C Morphology of the indicated paws from WT and KO mice on day 28. H&E staining (D) or Safranin O staining (E) of the knee joints from WT and KO mice on day 28 after the 2nd immunization. Scale bar = 200 μm. F ELISA analysis of the indicated cytokines in the sera of WT and KO mice after the 2nd immunization. G FACS of the percentages of IFN-γ-producing CD4+ T cells (Th1 cells) and IL-17a-producing CD4+ T cells (Th17 cells) in spleens from WT and KO mice after the 2nd immunization. **P < 0.01; ***P < 0.001; ****P < 0.0001. Two-way ANOVA (A), unpaired Student’s t test (B, F).
KDM5B deficiency protects mice from endotoxin shock through inhibiting macrophage-mediated inflammatory responses
Endotoxin shock models were performed subsequently to further investigate the pathological function of KDM5B in inflammatory responses in vivo. Intraperitoneal (i.p.) injection of endotoxin such as LPS and poly(I:C) into mice results in the activation of innate immune system and subsequent inflammatory tissue injury. KDM5B-KO mice produced much less pro-inflammatory cytokines including IL-1β, IL-6, IL-12p70, TNF-α and IFN-β in the sera upon the non-lethal dose of LPS or poly(I:C) treatment (Fig. 2A). Histological analysis by H&E staining showed that KDM5B deficiency protected mice from the inflammatory tissue injury with functional alveolar walls, less infiltration of inflammatory cells and reduced hemorrhage in lung tissues (Fig. 2B). KDM5B-KO mice showed pro-longed survival after i.p. injection of LPS with the lethal dose, compared with WT littermates that all died within 3 days (Fig. 2C upper). Notably, KDM5B-KO mice were completely insensitive to the relative low dose of LPS (12 mg/kg body weight) while 50% survival rate on day 5 was observed for WT mice (Fig. 2C lower). Additionally, the LPS-induced peritonitis model was constructed for evaluating the role of KDM5B in macrophage activation and recruitment [31, 32]. FACS analysis showed that KDM5B-KO mice had the rather reduced percentage and absolute number of F4/80+CD11b+ macrophage recruited to the peritoneal cavity (Fig. 2D, E). To address whether KDM5B regulated inflammatory responses mainly through myeloid cells such as macrophages, we performed bone marrow reconstruction experiments (Fig. 2F; Supplementary Fig. 3K). CD45.1 mice reconstituted with KDM5B-deficient bone marrow cells (KO → WT) produced less pro-inflammatory cytokines including IL-1β, IL-6 and TNF-α in the endotoxin shock model, compared with those reconstituted with WT bone marrow cells (WT → WT) (Fig. 2G). In addition, CD45.1 mice reconstituted with KDM5B-deficient bone marrow cells had less tissue injury and inflammatory cell infiltration in their lungs in contrast to control mice (Fig. 2H). The above in vivo data demonstrate that KDM5B is essential for the pro-inflammatory cytokine production in both acute and chronic inflammation model, and KDM5B deficiency protects mice from inflammation-associated tissue injury in the myeloid cell-intrinsic manner.
A ELISA analysis of the indicated cytokines in the sera of WT and KO mice 8 h after the intraperitoneal injection of LPS (12 mg/kg body weight) or Poly(I:C) (20 mg/kg body weight) (n = 6 mice per group). B H&E staining of lung tissues from WT and KO mice treated as in A. Scale bar = 200 μm. C Survival rates of WT and KO mice after the intraperitoneal injection of LPS (upper, 20 mg/kg body weight; lower, 12 mg/kg body weight) (n = 10-12 mice per group). D FACS of the percentages of F4/80+CD11b+ macrophages in peritoneal lavage fluids from WT and KO mice 8 h after the intraperitoneal injection of LPS (100 ng/per mouse) (n = 5 mice per group). E The percentage (left) and absolute number (right) of macrophages as in D. F The model of reconstitution of bone marrows from KDM5B-KO or WT mice (CD45.2) into CD45.1 WT mice. G ELISA analysis of the indicated cytokines in the sera of CD45.1 mice reconstituted with bone marrow cells from KDM5B-KO or WT mice 8 h after the intraperitoneal injection of LPS (12 mg/kg body weight) (n = 5 mice per group). H H&E staining of lung tissues from WT and KO mice as in G. Scale bar = 100 μm. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. One-way ANOVA (A), Log-rank test (C), unpaired Student’s t test (E, G).
Impaired TLR-triggered pro-inflammatory cytokine production in KDM5B-deficient macrophages
Considering that macrophages are the major mediator of TLR-triggered pro-inflammatory cytokine production in vivo, we next explored whether KDM5B influenced the development or differentiation of macrophages. FCAS analysis showed that the proportion and number of macrophages were comparable in both bone marrow and spleens between 8-week-old KO and WT mice (Supplementary Fig. 4A, B). Meanwhile, KDM5B had no effect on the percentage of peritoneal macrophages or in vitro differentiated BMDMs induced by M-CSF (Supplementary Fig. 4C, D). These data indicate that KDM5B does not influence the development and differentiation of macrophages. In response to LPS stimulation, KDM5B-deficient BMDMs exhibited lower mRNA levels of cytokines including IL-1β, IL-6, IL-12β, TNF-α and IFN-β (Fig. 3A). KDM5B deficiency also decreased mRNA levels of the above cytokines in BMDMs stimulated with poly(I:C) and Pam3CSK4 (Fig. 3B). KDM5B-deficient BMDMs produced less IL-6, TNF-α and IFN-β in the cell culture medium after treatment with LPS, poly(I:C) or Pam3CSK4 (Fig. 3C). Meanwhile, the lower protein level of pro-IL-1β was detected in KDM5B-KO BMDMs than that in WT BMDMs stimulated with LPS, poly(I:C) or Pam3CSK4 (Fig. 3D, E). We next investigated whether Kdm5b silencing affected cytokine production in the activated macrophages. The silencing of Kdm5b medicated by the specific siRNA significantly inhibited LPS-induced transcriptional expression of these pro-inflammatory cytokine such as Il1b, Il6, Il12b and Tnf (Supplementary Fig. 4E–G). These data demonstrate that KDM5B deficiency or silencing impairs the production of pro-inflammatory cytokines and type I interferon in macrophages upon different inflammatory stimuli.
A Q-PCR analysis of the mRNA levels of pro-inflammatory cytokines in WT and KDM5B-deficient (KO) BMDMs stimulated with LPS (100 ng/ml) for the indicated times. B Q-PCR analysis of the mRNA levels of pro-inflammatory cytokines in WT and KO BMDMs stimulated with LPS, poly(I:C) (pI:C) or Pam3CSK4 (Pam3) (100 ng/ml, 20 μg/ml and 1 μg/ml, respectively) for 6 h or left untreated (UT). C ELISA analysis of pro-inflammatory cytokines in culture supernatants of WT and KO BMDMs stimulated with LPS (100 ng/ml) or poly(I:C) (20 μg/ml) for 6 h. Immunoblot analysis of the indicated proteins in WT and KO BMDMs stimulated with LPS for the indicated times (D) or stimulated with the indicated ligands for 6 h (E). **P < 0.01; ***P < 0.001; ****P < 0.0001. One-way ANOVA (A–C).
KDM5B is required for NF-κB activation and transcriptional activity in macrophages
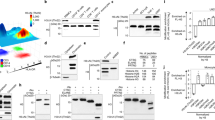
PAMPs were recognized by TLR receptors in macrophages and launched the different signaling cascades including MAPK and NF-κB pathway [33, 34]. Immunoblot assays showed that there was no substantial difference in the phosphorylation levels of ERK, JNK and p38 induced by LPS or Pam3CSK4 between KO and WT macrophages, suggesting that KDM5B had no effect on MAPK signaling activation (Fig. 4A, B). When it comes to NF-κB pathway, KDM5B did not affect the phosphorylation of IKKα/β. However, in KDM5B-deficient macrophages, the phosphorylation level of p65 was inhibited, which was might owing to the higher protein level of IκBα (Fig. 4A, B). Previous studies confirm that IκBα is the major negative regulator of NF-κB activation through inhibiting p65 phosphorylation and translocation into nucleus [35, 36]. Dual luciferase reporter gene assays of NF-κB further showed that KDM5B overexpression promoted both myeloid differentiation primary response gene 88 (MyD88)-induced and TNF receptor-associated factor 6 (TRAF6)-induced NF-κB activation in a dose-dependent manner, confirming that KDM5B facilitated NF-κB transcription activity (Fig. 4C). KDM5B was reported to remove promoter-associated H3K4me3 modification with both Jmjc and Jmjn domains, the two classic demethylase activity domains as other family members [37, 38]. To determine whether KDM5B regulated NF-κB activation dependent on its demethylase activity, we constructed two demethylase activity mutants of KDM5B, one lacking only Jmjc domain (called Jmjc-null) and the other lacking both Jmjc and Jmjn domains (called N terminal-null) (Fig. 4D). Interestingly, only full-length of KDM5B could promote MyD88-induced and TRAF6-induced NF-κB activation in HEK293T cells but two demethylase activity mutants of KDM5B did not (Fig. 4E). These data indicate that KDM5B is essential for TLR-triggered NF-κB activation dependent on its demethylase activity.
Immunoblot analysis of the indicated proteins in WT and KO BMDMs stimulated with LPS (100 ng/ml) (A) or Pam3CSK4 (1 μg/ml) (B) for the indicated times. C Dual luciferase reporter gene analysis of NF-κB-luciferase activity in HEK293T cells 24 h after transfection with NF-κB-luciferase reporter plasmid (NF-κB-luci), the indicated amounts of KDM5B-expressing plasmids and MyD88 (upper) or TRAF6 (lower). D Schematic diagram of the WT or mutants of KDM5B-expressing plasmids. E Dual luciferase receptor gene analysis of NF-κB-luciferase activity in HEK293T cells 24 h after transfection with NF-κB-luci, the indicated KDM5B mutants and MyD88 (left) or TRAF6 (right). **P < 0.01; ****P < 0.0001. One-way ANOVA (C, E).
KDM5B inhibitor GSK467 restrains NF-κB signaling activation and pro-inflammatory cytokine production in vitro and in vivo
Recent study identified a KDM5B enzyme activity inhibitor, GSK467, with a selective inhibition function on KDM5B but not KDM4C, KDM6 or other Jumonji family members [39]. Thus, we observed the effects of GSK467 on macrophage activation and inflammatory responses. GSK467 treatment (5 μM) resulted in the elevated H3K4me2 and H3K4me3 modification in macrophages, indicating GSK467 effectively inhibited KDM5B demethylase activity (Fig. 5A). GSK467 significantly suppressed the transcriptional induction of Il1b, Il6, Il12b and Tnf in macrophages upon LPS stimulation (Fig. 5B). In addition, the activation of LPS-induced NF-κB signaling cascade was also impaired in GSK467-treated macrophages with more IκBα protein expression and less phosphorylation level of p65 (Fig. 5C). These in vitro data confirmed the inhibitory function of GSK467 in NF-κB-triggered pro-inflammatory cytokine production. We next observed in vivo effects of GSK467 in endotoxin shock model. Firstly, we treated mice with different doses of GSK467 to determine an appropriate in vivo dosage without systemic toxicity. TUNEL staining of lung tissues showed that the low dose of GSK467 did not induce cell apoptosis in lung tissues except for mice from 20 mg/kg GSK467 group with a little proportion of TUNEL-positive cells (Supplementary Fig. 5A). Mice treated with 10 mg/kg or 20 mg/kg GSK467 produced much less IL-6 in sera in LPS-induced endotoxin shock model (Supplementary Fig. 5B). These data indicated that 10 mg/kg GSK467 contributed to the immunosuppression condition without toxicity. ELISA assays further showed that mice with GSK467 treatment (10 mg/kg) produced less pro-inflammatory cytokines in their sera 8 h after LPS challenge (Fig. 5D). Lung tissues from GSK467-treated mice exhibited more milder inflammatory injury with relatively intact alveolar structure and less infiltration of inflammatory cells (Fig. 5E). These above findings further confirm that KDM5B promotes inflammatory responses in vivo and in vitro in demethylase activity-dependent manner.
A Immunoblot analysis of the indicated proteins in BMDMs treated with GSK467 (5 μM) or DMSO overnight. Q-PCR analysis of the mRNA levels of pro-inflammatory cytokines (B) and immunoblot analysis of the indicated proteins (C) in BMDMs treated as in A followed by stimulation with LPS (100 ng/ml) for the indicated times. D ELISA analysis of pro-inflammatory cytokines in the sera of mice pre-treated with GSK467 (10 mg/kg per body weight) or DMSO followed by intraperitoneal injection with LPS (12 mg/kg per body weight) for 8 h (n = 5 per group). E H&E staining of lung tissues from mice treated as in D. **P < 0.01; ***P < 0.001; ****P < 0.0001. One-way ANOVA (B), unpaired Student’s t test (D).
KDM5B represses the transcriptional induction of Nfkbia gene
Since our data found that IκBα protein level in KDM5B-KO macrophages was much higher than that in WT macrophages, we further explore whether KDM5B influenced the transcription level of Nfkbia, the gene encoding IκBα. KDM5B deficiency markedly increased the mRNA level of Nfkbia in BMDMs stimulated with LPS, poly(I:C) or Pam3CSK4 (Fig. 6A, B). To confirm the selective regulatory function of KDM5B in Nfkbia mRNA expression, we also detected the expression of Nfkbib, Ikbka and Ikbkb, which respectively encoded IκBβ, IKKα and IKKβ, and found that KDM5B deficiency had no influence on these gene expression expect that of Nfkbia (Fig. 6C). Next, we analyzed the NFKBIA/Nfkbia expression in autoimmune diseases and found in synovial macrophages from RA patients, the level of NFKBIA was much higher than health control (Supplementary Fig. 6A). A more interesting thing was that there existed a strong negative correlation between the expression levels of KDM5B and NFKBIA in these samples of synovial macrophages (Supplementary Fig. 6B). In addition, NFKBIA was also highly expressed in the PBMCs from MS patients, which was also negatively correlated with the expression level of KDM5B (Supplementary Fig. 6C, D). We also compared the Nfkbia expression in colon tissues during different time period in a mouse experimental colitis model induced by DSS treatment, it turned out that Kdm5b expression was suppressed while Nfkbia level was elevated after DSS treatment, and there was a strong negative relationship between expression levels of these two genes (Supplementary Fig. 6E, F). Next, we analyzed the expression levels of KDM5B and NFKBIA in RA patients prior to or after anti-inflammatory or anti-rheumatic therapeutics. Current approved therapeutics for RA includes methotrexate (MTX), tocilizumab (TCZ) and rituximab (RTX), which target proliferating adaptive immune cells, interleukin-6 receptor (IL-6R) and CD20+ B cells respectively. Although these current approved therapeutics did not significantly change the expression of KDM5B and NFKBIA, the correlation analysis showed the significant negative relationship between KDM5B and NFKBIA expression levels (Supplementary Fig. 6G–I). These results suggest that in autoimmune disease settings, KDM5B may have an inhibitory role in NFKBIA mRNA expression.
Q-PCR analysis of the Nfkbia mRNA expression in WT and KO BMDMs stimulated with LPS (100 ng/ml) for the indicated times (A) or stimulated with the indicated ligands for 2 h (B). C Q-PCR analysis of the mRNA levels of the indicated genes in WT and KO BMDMs stimulated with LPS (100 ng/ml) for 1 h. D Dual luciferase reporter gene analysis of Nfkbia (−2000 to +121 region of the Nfkbia promoter) luciferase reporter activity in RAW264.7 cells transfected with the indicated amounts of mock vector and KDM5B-expressing plasmid followed by stimulation with LPS for 2 h or left untreated. E Dual luciferase reporter gene analysis of Nfkbia (IκBα), Nfkbib (IκBβ), Ikbka (IKKα) or Ikbkb (IKKβ) gene promoter-driven luciferase reporter activity in RAW264.7 cells transfected with mock vector or KDM5B-expressing plasmid followed by stimulation with LPS for 2 h. F Q-PCR analysis of Nfkbia mRNA expression in WT and KO BMDMs stimulated with LPS followed by treatment with ActD for the indicated times. G Immunoblot analysis of the indicated proteins in hMDMs transfected with Myc-tagged NFKBIA plasmid. H Immunoblot analysis of the indicated proteins in hMDMs treated as in G followed by stimulation with LPS (100 ng/ml) for the indicated times. I Q-PCR analysis of the mRNA levels of IL1B and IL6 genes in hMDMs treated as in G followed by stimulation with LPS (100 ng/ml) for 4 h. *P < 0.05, **P < 0.01; ***P < 0.001; ****P < 0.0001. One-way ANOVA (A, B, D, F, I), unpaired Student’s t test (C, E).
Dual luciferase reporter gene assay was next performed to confirm whether KDM5B could directly repress Nfkbia promoter-driven transcriptional activity. KDM5B overexpression decreased the reporter gene activity of the Nfkbia promoter but not Nfkbib, Ikbka or Ikbkb promoter in RAW264.7 cells stimulated with LPS (Fig. 6D, E). To exclude the post-transcriptional effect of KDM5B on Nfkbia expression, we treated LPS-activated KO and WT macrophages with actinomycin D (ActD) for transcriptional inhibition, and found no difference in mRNA destabilization of Nfkbia between KO and WT macrophages (Fig. 6F). To validate the effect of IκBα overexpression on human macrophage activation, human monocyte-derived macrophages (hMDMs) were cultured and transfected with Myc-tagged NFKBIA (Fig. 6G). IκBα overexpression was sufficient to inhibit LPS-triggered the activation of NF-κB signaling in macrophages, including the markedly impaired phosphorylation of p65 and the suppressed transcriptional induction of IL1B and IL6 (Fig. 6H, I). These data indicate that KDM5B plays an important role in the transcriptional repression of NFKBIA/Nfkbia gene in both human and mouse inflammatory macrophages.
KDM5B selectively binds to the Nfkbia promoter for erasing H3K4me3 modification and regulating chromatin accessibility
Given that KDM5B was reported as the transcriptional suppressor in various settings [40], we hypothesized that KDM5B might bound to the functional gene regions that were important in Nfkbia transcription. In this way, CUT&Tag followed by sequencing was done using KDM5B-specific antibody in BMDMs stimulated with LPS or not. Motif analysis by HOMER found that although two groups exhibited similar binding motif of KDM5B, there existed a quite different binding preference in LPS-activated macrophages (Fig. 7A). Next, we compared the target genes of KDM5B in macrophages stimulated with LPS or left untreated and found 252 LPS-induced binding genes (UP genes) and 88 LPS-repressed binding genes (DOWN genes) (Fig. 7B). With the gene functional annotation, we identified 13 immune-associated target genes (UP: 11 and DOWN: 2) (Fig. 7B; Supplementary Fig. 7A). Nfkbia gene was identified as the direct target of primary inflammatory signaling factor regulated by KDM5B in BMDMs stimulated with LPS, which was also confirmed by integrative genomics viewer (IGV) analysis (Fig. 7C). There existed two narrow peaks bound by KDM5B within the Nfkbia promoter, and motif enrichment analysis showed that transcription factor Sp1, NFAT and Nkx2.1 might be the downstream transcription factors influenced by KDM5B in LPS-stimulated macrophages (Supplementary Fig. 7B, C). A20, encoded by Tnfaip3, was also the widely-accepted negative regulator of NF-κB activation [41]. As the negative control, there was no specific binding peak in Tnfaip3 gene locus, or in pro-inflammatory cytokine Il6 gene locus (Supplementary Fig. 7D, E). Then we performed ChIP assays to verify the direct enrichment of KDM5B at the promoter of Nfkbia gene, which was increasingly induced by LPS treatment during the very early period in macrophages (Fig. 7D). However, there was no difference of KDM5B enrichment at the promoter of Tnfaip3 or Il6 between macrophages stimulated with LPS or left untreated (Fig. 7D). Considering the classical function of KDM5B in erasing the histone methylation modification of H3K4, we detected the H3K4me3 level at the promoter of Nfkbia gene, and observed the significant increase of H3K4me3 level in KDM5B-KO BMDMs (Fig. 7E). Notably, DNase I sensitivity assay showed that the chromatin accessibility in the Nfkbia promoter was increased in KDM5B-KO BMDMs after LPS stimulation, indicating that KDM5B deficiency affected the chromatin remodeling of the regulatory gene region around the transcription start site (TSS) of Nfkbia (Fig. 7F). In addition, the enrichment of RNA pol II in the promoter of Nfkbia was also elevated in KDM5B-KO BMDMs stimulated with LPS (Fig. 7G). The treatment of KDM5B inhibitor GSK467 also led to the increased H3K4me3 level, chromatin accessibility and RNA Pol II recruitment in the Nfkbia promoter of BMDMs (Supplementary Fig. 8A–C). To sum up, KDM5B is selectively recruited to the promoter of Nfkbia gene, removing H3K4me3 and inhibiting the chromatin accessibility for RNA Pol II-centered transcription machinery working (Supplementary Fig. 9).
A The motif analysis of KDM5B-bound peaks in BMDMs stimulated with LPS for 1 h or left untreated. B The strategy for identifying the target immune genes of KDM5B in macrophages. C IGV analysis of KDM5B (our work), H3K4me1 (GSE38377), H3K4me3 (GSE38377), ATAC (GSE108585) and RNA Pol II (GSE17631) signals in the gene locus of Nfkbia. D ChIP-qPCR analysis of KDM5B enrichment at the promoter of the indicated genes in BMDMs stimulated with LPS (100 ng/ml) for the indicated times. E ChIP-qPCR analysis of H3K4me3 enrichment at the promoter of Nfkbia gene in WT and KO BMDMs stimulated with LPS for 1 h. F DNase I sensitivity analysis of the Nfkbia promoter in the nuclei of LPS-stimulated WT and KO BMDMs. G ChIP-qPCR analysis of RNA Pol II enrichment at the promoter of Nfkbia gene in WT and KO BMDMs stimulated with LPS for 1 h or left untreated. *P < 0.05; **P < 0.01; ***P < 0.001. One-way ANOVA (D–G).
Discussion
In this work, we revealed the essential role of KDM5B in NF-κB activation and macrophage-mediated inflammatory responses in vivo and in vitro. Recently, a work by Liu and his colleges found that in drosophila, KDM5 family transcriptionally regulated component genes of the immune deficiency (IMD) signaling pathway and subsequent host-commensal bacteria homeostasis in a demethylase-dependent manner [42]. KDM5 inhibition also restrained the inhibitory effect of PPARγ agonist on IL-17A expression in Th17 responses [43]. Although KDM5 family removed the di/trimethylation of H3K4 in many cells, little work focuses their distinctive functions of different family members in immune regulation. Zhao et al. reported that KDM5A was required for NK cell activation by inhibiting suppressor of cytokine signaling 1 (SOCS1) expression. Mechanistically, KDM5A bound to the SOCS1 promoter in resting NK cells, leading to the elevated H3K4me3 modification and improved chromatin configuration for SOCS1 transcription [44]. In addition, several studies identified the regulatory role of KDM5 in antiviral immunity. For example, H3K4 demethylase KDM5B and KDM5C were responsible to repress STING transcription and KDM5 blockade triggered robust interferon responses in breast cancer cells [45]. Respiratory syncytial virus (RSV) infection induced the up-regulated expression of KDM5B in murine DCs, and KDM5B deficiency in DCs resulted in higher production of IFN-γ and reduced IL-4 and IL-5, indicating KDM5B could regulate Th2 cytokines when RSV infection [46]. However, our work found the down-regulation of KDM5B expression in the setting of multiple autoimmune diseases mainly owing to the transcription inhibition, which might be the protective manner of host immune to avoid excessive inflammatory responses and tissues injury. Nevertheless, the difference between KDM5B and other KDM5 family members were largely not clear.
Immune system and immune responses are generally in a precious equilibrium, involving kinds of regulatory mechanisms controlling the activation and inhibition of immune cell function, and participating in the initiation and resolution of inflammation. For example, the signal transduction of NF-κB is strictly regulated on different levels. A20 removes K63-linked poly-ubiquitin from NEMO after TNF stimulation, whose activity is promoted by IKK2-mediated phosphorylation of A20 at Ser381 [47, 48]. Kinase calcium/calmodulin-dependent protein kinase II (CaMKII) promotes NF-κB activation through directly binding and activating TAK1 in macrophages [49]. IκBα is the major player of negative feedback for turning-off the NF-κB response and maintains the moderate activation of NF-κB pathway by the coordinated degradation and synthesis of IκBα protein [50]. IκBα is degraded following exposure to inflammatory cytokines or microbial products and results in a decreased inhibitory effect on NF-κB, which is regulated by a variety of post-translational modifications including the phosphorylation at Ser32, Ser36 and Tyr42, ubiquitination at Lys21 and SUMOylation at Lys21 and Lys38 [51,52,53]. Recent works also demonstrated that the expression of IκBα was subjected to transcriptional regulation. The ChIP-seq of high-mobility group box1 (HMGB1) in human retinal endothelial cells identified NFKBIA as the binding site of HMGB1, which inhibited cell proliferation and promoted apoptosis [54]. Our previous work also showed that ZBTB20 inhibited IκBα gene transcription and promoted macrophage-mediated inflammatory responses [55]. In addition, Wang et al. revealed H3K4 methyltransferase Mixed lineage leukemia 1 (MLL1) was required for the increased H3K4 trimethylation level at the Nfkbia promoter induced by TNF-α [56]. So, whether KDM5B functioned as the co-repressor of these transcriptional factors including zinc finger and BTB domain-containing 20 (ZBTB20), or influenced the recruitment of activating transcriptional regulators such as MLL1 remained to be studied. Furthermore, beyond the NF-κB dogma, IκBα has been found to associate with unclear proteins such as HDACs or nuclear co-repressors, and in primary keratinocytes it bound the chromatin at specific genes and contributed to tumorigenesis [57, 58]. Here we found the strong negative correlation between KDM5B and NFKBIA expression in activated macrophages in the setting of autoimmune diseases. In this way, whether KDM5B-IκBα axis takes effects in other types of cells that lead to organ dysfunction or tumorigenesis is still largely unknown.
The regulatory role of histone methylation has been implicated in the development and function of immune cells, whose mutation or dysfunction might lead to immune deficiency or autoimmune diseases. For example, PBMCs from psoriasis patients exhibited reduced levels of histone H3/H4 acetylation and increased H3K4 methylation, and another work found reduced DNA methylation of gene including programmed cell death 1 ligand 2 (PDCD1LG2) in skin cells was related to immune activation and autoimmunity [59, 60]. We previously identified the histone demethylase KDM2B was indispensable for Il6 transcriptional induction through Brg1-mediated chromatin remodeling, not dependent on its H3K4me3 demethylase activity [61]. KDM5B deficiency was linked to the chromatin accessibility of Nfkbia gene regulatory regions, however, the epigenetic mechanisms underlying the dynamic change of chromatin structure were not elaborated. In the inducible transcription of Nfkbia gene, certain methyltransferases or epigenetic readers could participate in chromatin remodeling and promote transcriptional machinery function, while KDM5B might functioned as the negative feedback or a transcription brake for this process.
Finally, emerging therapies and perspective drugs were implicated in the treatment of autoimmune diseases like RA, including Tofacitinib, Baricitinib and other selective JAK inhibitors [62]. Meanwhile, monoclonal antibodies directed at pro-inflammatory cytokines such as Sirukumab and Tocilizumab were clinically proved to be effective. Although small-molecule inhibitors of epigenetic regulators were highlighted in enhancing anti-tumor immune responses, little had been applied to the treatment of autoimmune diseases [63]. Trichonstatin A (TSA) and suberoylanilide hydroxamic acid (SAHA) are two broad spectrum inhibitors of classical HDACs, directly binding to their active site for pharmacological inhibition. TSA-treated murine macrophages led to downregulation of a set of LPS-induced genes participating in multiple functions including inflammation, chemotaxis, antigen presentation and tissue repair [64]. Other researches also demonstrated that inhibition of HDACs had opposing effects. For example, Bode et al. and Halili et al. found that TSA and SAHA strongly inhibited the transcription of IL-12 p40 in DCs and macrophages respectively upon stimulation of TLRs [65, 66], while Zhang et al. showed that despite of the general inhibitory role of TSA in pro-inflammatory gene transcription shortly after LPS stimulation, the mRNA of Il6 was enhanced during the late phase in the presence of TSA [67]. A more recent work found that the dual-functional engagement of HDAC3 activity governed inflammatory responses in a gene-specific manner by SAHA administration, which provided the potential explanation for these contradictory functions. Given that the complexity of pharmacological effects, these epigenetic targets and inhibitors are being evaluated for therapy in inflammatory and autoimmune diseases [68]. In this work, we identified the therapeutic effects of KDM5B-specific inhibitor, GSK467 in LPS-induced systemic inflammation model with attenuation of pathogenic pro-inflammatory cytokine production in vivo. Nevertheless, the therapeutic results and the potential side-effects of GSK467 in autoimmune diseases such as RA, are expected to be gradually revealed. To sum up, we identify the protective mechanism of host immune system by inhibiting KDM5B expression and activity to avoid excessive inflammatory injury in autoimmune diseases.
Materials and methods
Reagents and antibodies
Complete Freund’s adjuvant (CFA) (F5881), Lipopolysaccharide (LPS) (Escherichia coli serotype 0111:B4), and poly(I:C) were purchased from Sigma-Aldrich. Pam3CSK4 was from Invivogen. Chicken Type II Collagen (20012) was purchased from Chondrex. ChIP-grade protein G magnetic beads (9006) and cell lysis buffer (9803) were from Cell Signaling Technology. Mouse IL-1β, IL-6, IL-12p70 and TNF-α ELISA kits were from R&D. Mouse IFN-β ELISA kits were from PBL. Antibodies against IL-1β (12242), Tri-Methyl-Histone H3 (Lys4) (C42D8), Di-Methyl-Histone H3 (Lys4) (C64G9), Histone H3 (1B1B2), phospho-ERK (9106), phospho-JNK (4668), phospho-p38 (9211), p65 (8242), phospho-p65 (3033), IκBα (112B2) (9247), IKKα (D3W6N) (61294), phosphor-IKKα/β (2697), and β-Actin (3700) were from Cell Signaling Technology. Antibody against KDM5B (A301-813) was from Bethyl Laboratories and Antibody against RNA Pol II (17-620) was from Millipore. Antibodies against CD45-BV605 (2D1), CD3-PerCP/Cy5.5 (17A2), CD19-APC (6D5), CD4-BV421 (GK1.5), CD8-BV510 (53-6.7), CD25-APC (PC61), PD-1-APC (29F.1A12), CXCR5-PE (J252D4), IFN-γ-APC (XMG1.2), IL-17a-PE (TC11-18H10.1), CD11b-BV510 (M1/70), and F4/80-FITC (QA17A29) were purchased from BioLegend. Antibody against FoxP3-PE (FJK-16s) was purchased from eBioscience.
Mice
Kdm5b-KO mice in C57BL/6 background were generated by CRISPR/Cas9 technology via a 428-base pair (bp) fragment deletion and frameshift mutation from the Kdm5b gene, resulting in the frameshift mutation and gene knockout. B6.SJL-Ptprca Pepcb (B6 CD45.1) (Strain# 002014) mice were purchased from the Jackson Laboratory. Wild type C57BL/6 mice (6–8 weeks old) were obtained from Joint Ventures Sipper BK Experimental Animal Company (Shanghai, China). Mice were bred in specific pathogen-free conditions. All animal experiments were undertaken in accordance with the National Institutes of Health’s Guide for the Care and Use of Laboratory Animals.
Clinical specimens
The peripheral blood mononuclear cells (PBMCs) of 38 subjects (28 RA cases and 10 health control) were recruited and collected from Changhai Hospital (Shanghai, China). Briefly, 10 ml anticoagulated whole blood was obtained and then PBMCs were collected by Ficoll gradient centrifugation. All the RA patients met the 2010 criteria of the American College of Rheumatology and the European Union League Against Rheumatism. Active RA was defined as having a 28-joint disease activity score (DAS28) of 2.6 or higher. For health control, subjects with severe cardiovascular diseases, liver and kidney dysfunction, malignant tumor and other autoimmune or musculoskeletal disorders were excluded. All samples were collected with written informed consent from the patients and health control.
Collagen-induced arthritis model
Eight-week-old mice were used to establish CIA models as previously described [69]. Bovine type II collagen emulsified in complete Freund’s adjuvant (CFA) was injected into mouse tail base on day 0 for the first immunization. Then a booster immunization was administered on day 21. We evaluated the severity of redness and swelling in the wrist and paw in a scale of 0–4. The maximum score is 16. All mice were scored blind to genotype. Knee joints were fixed in paraformaldehyde for further H&E and Safranin O staining. Sera were collected for cytokine measurement by enzyme-linked immunosorbnent assay (ELISA). Spleen was harvested for analyzing immune cell clustering via fluorescence-activated cell sorter (FACS).
LPS-induced systemic inflammation model
For non-lethal endotoxin shock model, 8-week-old mice were intraperitoneally injected with LPS (12 mg/kg body weight) [33]. Sera were collected 8 h after injection for cytokine measurement via ELISA. Lung tissues were examined by H&E staining. For lethal endotoxin shock model, 8-week-old mice were intraperitoneally injected with LPS (20 mg/kg body weight) and observed for survival rate, which was similar to previous study [55]. For peritonitis model, 8-week-old mice were intraperitoneally injected with LPS (100 ng/mouse) [31]. The peritoneal exudate cells were collected 8 h after injection and analyzed by FACS.
Cell culture
Murine bone marrow-derived macrophages (BMDMs) were prepared as previously described [61]. Human monocyte-derived macrophages (hMDMs) were collected from PBMCs of healthy volunteers and cultured by elutriation and GM-CSF-induced differentiation (50 ng/ml; Peprotech) as described before [5]. HEK293T cell lines and RAW264.7 cell lines were obtained from the American Type Culture Collection (ATCC), and cultured in DMEM medium or RPMI 1640 medium with 10% FBS (Gibco) as described [70].
Flow cytometry and intracellular cytokine staining
Single cell suspensions were prepared from the indicated tissues or peritoneal cavity and subjected to FACS using SONY ID7000. FACS data were analyzed by FlowJo software. Cells were stained with fluorochrome-labeled antibodies after incubation with anti-CD16/CD32 to block non-specific antibody binding. For intracellular cytokine detection of IFN-γ and IL-17a, cells were stimulated with Cell Activation Cocktail (with Brefeldin A) (BioLegend) for 4–6 h. After incubation, cells were stained with CD16/32 and other antibodies against surface molecules. After surface staining, cells were fixed and permeabilized with the fixation/permeabilization kit (BD Biosciences) and stained for intracellular cytokines. Intracellular FoxP3 staining was performed using Foxp3/Transcription Factor Staining Buffer Set (Invitrogen) without cell stimulation.
RNA isolation and quantitative-PCR (Q-PCR) assays
Total RNA was extracted from cells using TRIzol reagent (ThermoFisher Scientific) according the instruction. Q-PCR assays were performed using a LightCycler 480 (Roche) and SYBR RT-PCR kit (Takara). Data were normalized to Actb expression.
Immunoblot analysis
Immunoblot analysis was performed as described previously [71, 72]. Briefly, total proteins from cells were extracted with cell lysis buffer (Cell Signaling Technology) with addition of a protease inhibitor cocktail and PMSF (Merck). The protein concentration was measured with a BCA protein assay kit (ThermoFisher Scientific) and subject to immunoblot assays using BIO-RAD electrophoresis system.
Plasmid construction
Vectors encoding mouse Kdm5b (NM_152895.2) and human NFKBIA (NM_020529.3) were constructed by PCR-based amplification from cDNA of mouse macrophages or hDMDs and then cloned into the pcDNA3.1-V5-His or pcDNA3.1-Myc eukaryotic expression vector (Invitrogen) as described [61]. Mutants of KDM5B were constructed by PCR-based amplification from the wild type plasmid and the primers used were listed as follows: KDM5B-WT (Forward: 5′-GGGGTACCGTGATGGAGCCGGCCA CCACGCTGCCCCCAG-3′; Reverse: 5′-GCYCYAGACTCTTTCGGCTTGGTGCGT CCTTCCCAGTAC-3′), Jmjc-null (Forward: 5′-CCTAAGTGTTTGGCTCAAGAATG TAATGTTGATTGGCTGCC-3′; Reverse: 5’-GGCAGCCAATCAACATTACATTCTT GAGCCAAACACTTAGG-3′), N terminal-null (Forward: 5′-GGGGTACCGTGATGC TGGATGATCTCTATCCAATGATG-3′; Reverse: 5′- GCYCYAGACTCTTTCGGCTT GGTGCGTCCTTCCCAGTAC-3′).
RNA interference and plasmid transfection
SiRNA targeting Kdm5b (siGENOME SMART pool siRNA M-058167-01) and control siRNA (siGENOME non-targeting siRNA Pool #1) were from Dharmacon. The siRNA duplexes were transfected into macrophages using Lipofectamine RNAiMax (ThermoFisher Scientific) according to the manufacturer’s protocol. The indicated plasmids were transfected into HEK293T cells or RAW264.7 cells using jetPEI (Polyplus-transfection) or FuGene HD (Promega) respectively.
Dual luciferase reporter gene assay
HEK293T cells or RAW264.7 cells were co-transfected with a mixture of the indicated luciferase reporter plasmids, pRL-TK-Renukka-luciferase plasmid and KDM5B wild type or mutant plasmids. After 24–36 h, luciferase activities were measured using the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Data are normalized for transfection efficiency by dividing Firefly luciferase activity with the activity of Renilla luciferase.
Chromatin immunoprecipitation (ChIP)
BMDMs were firstly stimulated with LPS for the indicated times and cross-linked with 1% (v/v) methanol-free formaldehyde for 10 min, terminated with glycine solution, and then subjected to ChIP assays using the ChIP Assay Kit (Beyotime). The antibodies against KDM5B (2 μL/sample) and RNA Pol II (1 μL/sample) were from Bethyl Laboratories and Millipore respectively. Purified DNA was detected by Q-PCR and then normalized to the input DNA for each sample. The primers used for amplification and quantification in ChIP-qPCR were listed as follows: Nfkbia (Forward: 5′-AAAGTTCCCTGTGCATGACC-3′; Reverse: 5′-CTGGCAGGGGATTTCTCAG-3′), Tnfaip3 (Forward: 5′-TTGAATGGTGGTGGTCTTCA-3′; Reverse: 5′-TGAGGAGGAGGGGAATAACC-3′) and Il6 (Forward: 5’-CCTGCGTTTAAATAACATCAGCTTTG CTT-3’; Reverse: 5’-GCACAATGTGACGTCGTTTAGCATCGAA-3’).
Cleavage under targets & tagmentation (CUT&Tag)
CUT&Tag was performed using Hyperactive Universal CUT&Tag Assay Kit for Illumina (Vazyme, Shanghai) according to the product instructions. Briefly, BMDMs were stimulated with LPS and then harvested after PBS wash. Cells (1 × 105) were bound with concanavalin A-coated magnetic beads and then incubated with antibody against KDM5B (Bethyl Laboratories) at 4 °C overnight. On next day, samples were incubated with second antibody and then with Hyperactive pA/G-Transposon for 1 h. DNA was treated with TTBL at 37 °C for 1 h and extracted, following by purification for library amplification. The Next Generation Sequencing (second-generation high-throughput sequencing) technique was applied to sequence DNA. Genome mapping was performed using Bowtie (version: 0.12.8) and clean reads were compared to reference genome mm10. MACS (version 2.2.7.1) was applied to call peak. The bam file and R package DiffBind were used to analyze the differences between the peaks of the specified two samples. The default parameter was set as fold change ≥ 2 or ≤ 1/2 and P value < 0.05, and the differences between the obtained peaks were annotated. The annotation program was the same as the above peak annotation program, including different peak location information, annotation information, etc. The motif analysis was conducted using “HOMER” tool. And the result of HOMER motif is the motif information predicted based on the existing database within the peak. The findmotifsGenome.pl tool for HOMER was used to analyze the specific location of Motif at peak. The sequencing data are available in the GEO database under accession number GSE212808.
DNase I sensitivity assays
DNase I sensitivity assays were performed as described before [61]. Briefly, nuclei of BMDMs stimulated with LPS for 1 h were extracted by nuclear extraction Kit (Biotechnology, Shanghai) and then treated with DNase I (0.1 U/μL, Promega) at 37 °C for 30 min followed by termination with EDTA (50 mM). Genomic DNA was extracted and purified using QIAamp DNA micro kit (QIAGEN) and subjected to Q-PCR assays.
Statistical analysis
The statistical significance of differences between the two groups was determined by unpaired Student’s t-test. For comparison of more than two groups, one- or two-way ANOVA with the least significant difference t-test was performed. The statistical significance of survival curves was estimated according to the method of Kaplan and Meier, and curves were compared with the generalized Wilcoxon test. P values of less than 0.05 were considered statistically significant. The statistical tests were justified as appropriate according to assessment of normality and variance of the distribution of the data. No randomization or exclusion of data points was used.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Data supporting the present study are available from the corresponding author upon reasonable request. Original images of unprocessed immunoblot and tissue staining are available at Supplementary information.
References
Na YR, Stakenborg M, Seok SH, Matteoli G. Macrophages in intestinal inflammation and resolution: a potential therapeutic target in IBD. Nat Rev Gastroenterol Hepatol. 2019;16:531–43.
Watanabe S, Alexander M, Misharin AV, Budinger GRS. The role of macrophages in the resolution of inflammation. J Clin Investig. 2019;129:2619–28.
Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: a cell biological perspective. Annu Rev Immunol. 2015;33:257–90.
Yao X, Huang J, Zhong H, Shen N, Faggioni R, Fung M, et al. Targeting interleukin-6 in inflammatory autoimmune diseases and cancers. Pharmacol Ther. 2014;141:125–39.
Krausgruber T, Blazek K, Smallie T, Alzabin S, Lockstone H, Sahgal N, et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nat Immunol. 2011;12:231–8.
Udalova IA, Mantovani A, Feldmann M. Macrophage heterogeneity in the context of rheumatoid arthritis. Nat Rev Rheumatol. 2016;12:472–85.
Ross EA, Devitt A, Johnson JR. Macrophages: the good, the bad, and the gluttony. Front Immunol. 2021;12:708186.
Tardito S, Martinelli G, Soldano S, Paolino S, Pacini G, Patane M, et al. Macrophage M1/M2 polarization and rheumatoid arthritis: a systematic review. Autoimmun Rev. 2019;18:102397.
Daskalaki MG, Tsatsanis C, Kampranis SC. Histone methylation and acetylation in macrophages as a mechanism for regulation of inflammatory responses. J Cell Physiol. 2018;233:6495–507.
Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, et al. Metabolic regulation of gene expression by histone lactylation. Nature. 2019;574:575–80.
Lauterbach MA, Hanke JE, Serefidou M, Mangan MSJ, Kolbe CC, Hess T, et al. Toll-like receptor signaling rewires macrophage metabolism and promotes histone acetylation via ATP-citrate lyase. Immunity. 2019;51:997–1011.
Xhabija B, Kidder BL. KDM5B is a master regulator of the H3K4-methylome in stem cells, development and cancer. Semin Cancer Biol. 2019;57:79–85.
Yan G, Li S, Yue M, Li C, Kang Z. Lysine demethylase 5B suppresses CC chemokine ligand 14 to promote progression of colorectal cancer through the Wnt/beta-catenin pathway. Life Sci. 2021;264:118726.
Wong PP, Miranda F, Chan KV, Berlato C, Hurst HC, Scibetta AG. Histone demethylase KDM5B collaborates with TFAP2C and Myc to repress the cell cycle inhibitor p21(cip) (CDKN1A). Mol Cell Biol. 2012;32:1633–44.
Wang X, Gu M, Ju Y, Zhou J. Overcoming radio-resistance in esophageal squamous cell carcinoma via hypermethylation of PIK3C3 promoter region mediated by KDM5B loss. J Radiat Res. 2022;63:331–41.
Zhang SM, Cai WL, Liu X, Thakral D, Luo J, Chan LH, et al. KDM5B promotes immune evasion by recruiting SETDB1 to silence retroelements. Nature. 2021;598:682–7.
Kang K, Park SH, Chen J, Qiao Y, Giannopoulou E, Berg K, et al. Interferon-gamma represses M2 gene expression in human macrophages by disassembling enhancers bound by the transcription factor MAF. Immunity. 2017;47:235–50.
Kemppinen AK, Kaprio J, Palotie A, Saarela J. Systematic review of genome-wide expression studies in multiple sclerosis. BMJ Open. 2011;1:e000053.
Burczynski ME, Peterson RL, Twine NC, Zuberek KA, Brodeur BJ, Casciotti L, et al. Molecular classification of Crohn’s disease and ulcerative colitis patients using transcriptional profiles in peripheral blood mononuclear cells. J Mol Diagn. 2006;8:51–61.
Olsen J, Gerds TA, Seidelin JB, Csillag C, Bjerrum JT, Troelsen JT, et al. Diagnosis of ulcerative colitis before onset of inflammation by multivariate modeling of genome-wide gene expression data. Inflamm Bowel Dis. 2009;15:1032–8.
Stojanov S, Lapidus S, Chitkara P, Feder H, Salazar JC, Fleisher TA, et al. Periodic fever, aphthous stomatitis, pharyngitis, and adenitis (PFAPA) is a disorder of innate immunity and Th1 activation responsive to IL-1 blockade. Proc Natl Acad Sci USA. 2011;108:7148–53.
Aletaha D, Neogi T, Silman AJ, Funovits J, Felson DT, Bingham CO 3rd, et al. 2010 rheumatoid arthritis classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann Rheum Dis. 2010;69:1580–8.
Leyva-Illades D, Cherla RP, Galindo CL, Chopra AK, Tesh VL. Global transcriptional response of macrophage-like THP-1 cells to Shiga toxin type 1. Infect Immun. 2010;78:2454–65.
Martinez FO, Helming L, Milde R, Varin A, Melgert BN, Draijer C, et al. Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood. 2013;121:e57–69.
Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–23.
Cuartero S, Weiss FD, Dharmalingam G, Guo Y, Ing-Simmons E, Masella S, et al. Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nat Immunol. 2018;19:932–41.
Albert M, Schmitz SU, Kooistra SM, Malatesta M, Morales Torres C, Rekling JC, et al. The histone demethylase Jarid1b ensures faithful mouse development by protecting developmental genes from aberrant H3K4me3. PLoS Genet. 2013;9:e1003461.
Backe MB, Jin C, Andreone L, Sankar A, Agger K, Helin K, et al. The lysine demethylase KDM5B regulates islet function and glucose homeostasis. J Diabetes Res. 2019;2019:5451038.
Brand DD, Latham KA, Rosloniec EF. Collagen-induced arthritis. Nat Protoc. 2007;2:1269–75.
Teng MW, Bowman EP, McElwee JJ, Smyth MJ, Casanova JL, Cooper AM, et al. IL-12 and IL-23 cytokines: from discovery to targeted therapies for immune-mediated inflammatory diseases. Nat Med. 2015;21:719–29.
De Filippo K, Henderson RB, Laschinger M, Hogg N. Neutrophil chemokines KC and macrophage-inflammatory protein-2 are newly synthesized by tissue macrophages using distinct TLR signaling pathways. J Immunol. 2008;180:4308–15.
Yu L, Zhang B, Deochand D, Sacta MA, Coppo M, Shang Y, et al. Negative elongation factor complex enables macrophage inflammatory responses by controlling anti-inflammatory gene expression. Nat Commun. 2020;11:2286.
Liu X, Zhang P, Zhang Y, Wang Z, Xu S, Li Y, et al. Glycolipid iGb3 feedback amplifies innate immune responses via CD1d reverse signaling. Cell Res. 2019;29:42–53.
Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-kappaB signaling pathways. Nat Immunol. 2011;12:695–708.
Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–34.
Viatour P, Merville MP, Bours V, Chariot A. Phosphorylation of NF-kappaB and IkappaB proteins: implications in cancer and inflammation. Trends Biochem Sci. 2005;30:43–52.
Fu YD, Huang MJ, Guo JW, You YZ, Liu HM, Huang LH, et al. Targeting histone demethylase KDM5B for cancer treatment. Eur J Med Chem. 2020;208:112760.
Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z, et al. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc Natl Acad Sci USA. 2007;104:19226–31.
Johansson C, Velupillai S, Tumber A, Szykowska A, Hookway ES, Nowak RP, et al. Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat Chem Biol. 2016;12:539–45.
Zheng YC, Chang J, Wang LC, Ren HM, Pang JR, Liu HM. Lysine demethylase 5B (KDM5B): a potential anti-cancer drug target. Eur J Med Chem. 2019;161:131–40.
Vereecke L, Beyaert R, van Loo G. The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology. Trends Immunol. 2009;30:383–91.
Chen K, Luan X, Liu Q, Wang J, Chang X, Snijders AM, et al. Drosophila histone demethylase KDM5 regulates social behavior through immune control and gut microbiota maintenance. Cell Host Microbe. 2019;25:537–52.
Miao Y, Zheng Y, Geng Y, Yang L, Cao N, Dai Y, et al. The role of GLS1-mediated glutaminolysis/2-HG/H3K4me3 and GSH/ROS signals in Th17 responses counteracted by PPARgamma agonists. Theranostics. 2021;11:4531–48.
Zhao D, Zhang Q, Liu Y, Li X, Zhao K, Ding Y, et al. H3K4me3 demethylase Kdm5a is required for NK cell activation by associating with p50 to suppress SOCS1. Cell Rep. 2016;15:288–99.
Wu L, Cao J, Cai WL, Lang SM, Horton JR, Jansen DJ, et al. KDM5 histone demethylases repress immune response via suppression of STING. PLoS Biol. 2018;16:e2006134.
Ptaschinski C, Mukherjee S, Moore ML, Albert M, Helin K, Kunkel SL, et al. RSV-induced H3K4 demethylase KDM5B leads to regulation of dendritic cell-derived innate cytokines and exacerbates pathogenesis in vivo. PLoS Pathog. 2015;11:e1004978.
Wertz IE, Newton K, Seshasayee D, Kusam S, Lam C, Zhang J, et al. Phosphorylation and linear ubiquitin direct A20 inhibition of inflammation. Nature. 2015;528:370–5.
Mauro C, Pacifico F, Lavorgna A, Mellone S, Iannetti A, Acquaviva R, et al. ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NF-kappaB. J Biol Chem. 2006;281:18482–8.
Liu X, Yao M, Li N, Wang C, Zheng Y, Cao X. CaMKII promotes TLR-triggered proinflammatory cytokine and type I interferon production by directly binding and activating TAK1 and IRF3 in macrophages. Blood. 2008;112:4961–70.
Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–5.
Wang X, Peng H, Huang Y, Kong W, Cui Q, Du J, et al. Post-translational modifications of IkappaBalpha: the state of the art. Front Cell Dev Biol. 2020;8:574706.
Hendriks IA, Lyon D, Young C, Jensen LJ, Vertegaal AC, Nielsen ML. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat Struct Mol Biol. 2017;24:325–36.
Komives EA. Consequences of fuzziness in the NFkappaB/IkappaBalpha interaction. Adv Exp Med Biol. 2012;725:74–85.
Liang WJ, Yang HW, Liu HN, Qian W, Chen XL. HMGB1 upregulates NF-kB by inhibiting IKB-alpha and associates with diabetic retinopathy. Life Sci. 2020;241:117146.
Liu X, Zhang P, Bao Y, Han Y, Wang Y, Zhang Q, et al. Zinc finger protein ZBTB20 promotes Toll-like receptor-triggered innate immune responses by repressing IkappaBalpha gene transcription. Proc Natl Acad Sci USA. 2013;110:11097–102.
Wang X, Zhu K, Li S, Liao Y, Du R, Zhang X, et al. MLL1, a H3K4 methyltransferase, regulates the TNFalpha-stimulated activation of genes downstream of NF-kappaB. J Cell Sci. 2012;125:4058–66.
Mulero MC, Bigas A, Espinosa L. IkappaBalpha beyond the NF-kB dogma. Oncotarget. 2013;4:1550–1.
Aguilera C, Hoya-Arias R, Haegeman G, Espinosa L, Bigas A. Recruitment of IkappaBalpha to the hes1 promoter is associated with transcriptional repression. Proc Natl Acad Sci USA. 2004;101:16537–42.
Roberson ED, Liu Y, Ryan C, Joyce CE, Duan S, Cao L, et al. A subset of methylated CpG sites differentiate psoriatic from normal skin. J Investig Dermatol. 2012;132:583–92.
Ovejero-Benito MC, Reolid A, Sanchez-Jimenez P, Saiz-Rodriguez M, Munoz-Aceituno E, Llamas-Velasco M, et al. Histone modifications associated with biological drug response in moderate-to-severe psoriasis. Exp Dermatol. 2018;27:1361–71.
Zhou Q, Zhang Y, Wang B, Zhou W, Bi Y, Huai W, et al. KDM2B promotes IL-6 production and inflammatory responses through Brg1-mediated chromatin remodeling. Cell Mol Immunol. 2020;17:834–42.
Ferro F, Elefante E, Luciano N, Talarico R, Todoerti M. One year in review 2017: novelties in the treatment of rheumatoid arthritis. Clin Exp Rheumatol. 2017;35:721–34.
Hogg SJ, Beavis PA, Dawson MA, Johnstone RW. Targeting the epigenetic regulation of antitumour immunity. Nat Rev Drug Discov. 2020;19:776–800.
Serrat N, Sebastian C, Pereira-Lopes S, Valverde-Estrella L, Lloberas J, Celada A. The response of secondary genes to lipopolysaccharides in macrophages depends on histone deacetylase and phosphorylation of C/EBPbeta. J Immunol. 2014;192:418–26.
Bode KA, Schroder K, Hume DA, Ravasi T, Heeg K, Sweet MJ, et al. Histone deacetylase inhibitors decrease Toll-like receptor-mediated activation of proinflammatory gene expression by impairing transcription factor recruitment. Immunology. 2007;122:596–606.
Halili MA, Andrews MR, Labzin LI, Schroder K, Matthias G, Cao C, et al. Differential effects of selective HDAC inhibitors on macrophage inflammatory responses to the Toll-like receptor 4 agonist LPS. J Leukoc Biol. 2010;87:1103–14.
Zhang Q, Zhao K, Shen Q, Han Y, Gu Y, Li X, et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 2015;525:389–93.
Nguyen HCB, Adlanmerini M, Hauck AK, Lazar MA. Dichotomous engagement of HDAC3 activity governs inflammatory responses. Nature. 2020;584:286–90.
Xia M, Liu J, Wu X, Liu S, Li G, Han C, et al. Histone methyltransferase Ash1l suppresses interleukin-6 production and inflammatory autoimmune diseases by inducing the ubiquitin-editing enzyme A20. Immunity. 2013;39:470–81.
Huai W, Liu X, Wang C, Zhang Y, Chen X, Chen X, et al. KAT8 selectively inhibits antiviral immunity by acetylating IRF3. J Exp Med. 2019;216:772–85.
Liu X, Zhan Z, Li D, Xu L, Ma F, Zhang P, et al. Intracellular MHC class II molecules promote TLR-triggered innate immune responses by maintaining activation of the kinase Btk. Nat Immunol. 2011;12:416–24.
Wang Y, Wang P, Zhang Y, Xu J, Li Z, Li Z, et al. Decreased expression of the host long-noncoding RNA-GM facilitates viral escape by inhibiting the kinase activity TBK1 via S-glutathionylation. Immunity. 2020;53:1168–81.
Funding
This work was supported by the National Key R&D Program of China (2019YFA0801502), the National Natural Science Foundation of China (82071790, 82070415, 82271797), the Shuguang Program sponsored by the Shanghai Education Development Foundation and Shanghai Municipal Education Commission (18SG33, 19SG17), and the China National Postdoctoral Program for Innovative Talents (BX2021046).
Author information
Authors and Affiliations
Contributions
YZ, YG, YJ, YD, HC and YX performed the experiments, analyzed the data, and interpreted the results. YZ drafted the manuscript. XL and ZZ conceived the idea, designed the experiments, assisted data analyses, revised the manuscript, and provided funding for this study. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
All experiments with animals were approved by the Animal Ethics Committee of Naval Medical University. All studies involving human PBMC samples were approved by the Medical Ethics Committee of Naval Medical University.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Y., Gao, Y., Jiang, Y. et al. Histone demethylase KDM5B licenses macrophage-mediated inflammatory responses by repressing Nfkbia transcription. Cell Death Differ 30, 1279–1292 (2023). https://doi.org/10.1038/s41418-023-01136-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41418-023-01136-x