Abstract
Background
Molecular subtyping of bladder cancer has revealed luminal tumors generally have a more favourable prognosis. However, some aggressive forms of variant histology, including micropapillary, are often classified luminal. In previous work, we found long non-coding RNA (lncRNA) expression profiles could identify a subgroup of luminal bladder tumors with less aggressive biology and better outcomes.
Objective
In the present study, we aimed to investigate whether lncRNA expression profiles could identify high-grade T1 micropapillary bladder cancer with differential outcome.
Design, setting, and participants
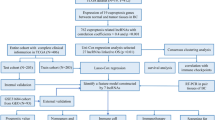
LncRNAs were quantified from RNA-seq data from a HGT1 bladder cancer cohort that was enriched for primary micropapillary cases (15/84). Unsupervised consensus clustering of variant lncRNAs identified a three-cluster solution, which was further characterised using a panel of micropapillary-associated biomarkers, molecular subtypes, gene signatures, and survival analysis. A single-sample genomic signature was trained using lasso-penalized logistic regression to classify micropapillary-like gene-expression, as characterised by lncRNA clustering. The genomic classifier (GC) was tested on luminal tumors derived from the TCGA cohort (N = 202).
Outcome measurements and statistical analysis
Patient and tumor characteristics were compared between subgroups by using X2 tests and two-sided Wilcoxon rank-sum tests. Primary endpoints were overall, progression-free and high-grade recurrence-free survival, calculated as the date of high-grade T1 disease at TURBT till date of death from any cause, progression, or recurrence, respectively. Survival rates were estimated using weighted Kaplan–Meier (KM) curves.
Results and limitations
Primary micropapillary HGT1 showed decreased FGFR3, SHH, and p53 pathway activity relative to tumors with conventional urothelial carcinoma. Many bladder cancer-associated lncRNAs were downregulated in micropapillary tumors, including UCA1, LINC00152, and MALAT1. Unsupervised consensus clustering resulted in a lncRNA cluster 1 (LC1) with worse prognosis that was enriched for primary micropapillary histology and the Luminal Unstable (LumU) molecular subtype. Interestingly, LC1 appeared to better identify aggressive HGT1 disease, compared to stratifying outcomes using primary histologic characteristics. A signature trained to identify LC1 cases showed good performance in the testing cohort, identifying seven cases with significantly worse survival (p < 0.001). Limitations include the retrospective nature of the study and the lack of a validation cohort.
Conclusions
Using the lncRNA transcriptome we identified a subgroup of aggressive HGT1 bladder cancer that was enriched with micropapillary histology. These data suggest that lncRNAs can facilitate the identification of aggressive micropapillary-like tumors, potentially improving patient management.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 24 print issues and online access
$259.00 per year
only $10.79 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Gene expression data of the T1 cohort have been uploaded to GEO (GEO: GSE136401). The TCGA RNA-seq data was available as part of the TCGA research network (http://cancergenome.nih.gov/).
References
Abufaraj M, Foerster B, Schernhammer E, Moschini M, Kimura S, Hassler MR, et al. Micropapillary urothelial carcinoma of the bladder: A systematic review and meta-analysis of disease characteristics and treatment outcomes. Eur Urol. 2019;75:649–58.
Witjes JA, Babjuk M, Bellmunt J, Bruins HM, De Reijke TM, De Santis M, et al. EAU-ESMO consensus statements on the management of advanced and variant bladder cancer—an international collaborative multistakeholder Effort (dagger): Under the auspices of the EAU-ESMO Guidelines Committees. Eur Urol. 2020;77:223–50.
Guo CC, Dadhania V, Zhang L, Majewski T, Bondaruk J, Sykulski M, et al. Gene expression profile of the clinically aggressive micropapillary variant of bladder cancer. Eur Urol. 2016;70:611–20.
Bowden M, Nadal R, Zhou CW, Werner L, Barletta J, Juanpere N, et al. Transcriptomic analysis of micropapillary high grade T1 urothelial bladder cancer. Sci Rep. 2020;10:20135.
Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25:152–65.
Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive molecular characterization of muscle-invasive bladder. Cancer Cell 2017;171:540–56.e25.
Necchi A, Raggi D, Gallina A, Ross JS, Fare E, Giannatempo P, et al. Impact of molecular subtyping and immune infiltration on pathological response and outcome following neoadjuvant pembrolizumab in muscle-invasive bladder cancer. Eur Urol. 2020;77:701–10.
Seiler R, Ashab HAD, Erho N, van Rhijn BWG, Winters B, Douglas J, et al. Impact of molecular subtypes in muscle-invasive bladder cancer on predicting response and survival after neoadjuvant chemotherapy. Eur Urol. 2017;72:544–54.
Sjodahl G, Eriksson P, Liedberg F, Hoglund M. Molecular classification of urothelial carcinoma: global mRNA classification versus tumour-cell phenotype classification. J Pathol. 2017;242:113–25.
Kamoun A, de Reynies A, Allory Y, Sjodahl G, Robertson AG, Seiler R, et al. A consensus molecular classification of muscle-invasive bladder cancer. Eur Urol. 2020;77:420–33.
Robertson AG, Groeneveld CS, Jordan B, Lin X, McLaughlin KA, Das A, et al. Identification of differential tumor subtypes of T1 bladder cancer. Eur Urol. 2020;78:533–7.
Lindskrog SV, Prip F, Lamy P, Taber A, Groeneveld CS, Birkenkamp-Demtroder K, et al. An integrated multi-omics analysis identifies prognostic molecular subtypes of non-muscle-invasive bladder cancer. Nat Commun. 2021;12:2301.
de Jong JJ, Zwarthoff EC. Molecular and clinical heterogeneity within the luminal subtype. Nat Rev Urol. 2020;17:69–70.
Gutschner T, Diederichs S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012;9:703–19.
Gibb EA, Vucic EA, Enfield KS, Stewart GL, Lonergan KM, Kennett JY, et al. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011;6:e25915.
Nguyen Q, Carninci P. Expression specificity of disease-associated lncRNAs: Toward personalized medicine. Curr Top Microbiol Immunol. 2016;394:237–58.
de Jong JJ, Liu Y, Robertson AG, Seiler R, Groeneveld CS, van der Heijden MS, et al. Long non-coding RNAs identify a subset of luminal muscle-invasive bladder cancer patients with favorable prognosis. Genome Med. 2019;11:60.
Ghafouri-Fard S, Taheri M. UCA1 long non-coding RNA: An update on its roles in malignant behavior of cancers. Biomed Pharmacother. 2019;120:109459.
Xian-Li T, Hong L, Hong Z, Yuan L, Jun-Yong D, Peng X, et al. Higher expression of Linc00152 promotes bladder cancer proliferation and metastasis by activating the Wnt/beta-catenin signaling pathway. Med Sci Monit. 2019;25:3221–30.
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F, et al. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin Cancer Res. 2014;20:1531–41.
Yang Y, Kaimakliotis HZ, Williamson SR, Koch MO, Huang K, Barboza MP, et al. Micropapillary urothelial carcinoma of urinary bladder displays immunophenotypic features of luminal and p53-like subtypes and is not a variant of adenocarcinoma. Urol Oncol. 2020;38:449–58.
Zinnall U, Weyerer V, Comperat E, Camparo P, Gaisa NT, Knuechel-Clarke R, et al. Micropapillary urothelial carcinoma: Evaluation of HER2 status and immunohistochemical characterization of the molecular subtype. Hum Pathol. 2018;80:55–64.
Batista da Costa J, Gibb EA, Bivalacqua TJ, Liu Y, Oo HZ, Miyamoto DT, et al. Molecular characterization of neuroendocrine-like bladder cancer. Clin Cancer Res. 2019;25:3908–20.
Grivas P, Bismar TA, Alva AS, Huang HC, Liu Y, Seiler R, et al. Validation of a neuroendocrine-like classifier confirms poor outcomes in patients with bladder cancer treated with cisplatin-based neoadjuvant chemotherapy. Urol Oncol. 2020;38:262–8.
Genitsch V, Kollar A, Vandekerkhove G, Blarer J, Furrer M, Annala M, et al. Morphologic and genomic characterization of urothelial to sarcomatoid transition in muscle-invasive bladder cancer. Urol Oncol. 2019;37:826–36.
Author information
Authors and Affiliations
Contributions
JJdJ, EAG, and JB designed the study. JJdJ performed gene expression analyses and classifier development, with suggestions from EAG and JB. JJdJ, EAG, and JB wrote the manuscript with feedback from all authors. JJdJ, EAG., JB, JP, and MA curated data for the study. All authors provided critical review and revisions, and all authors approved the final version of the manuscript for submission and publication. EAG and JB contributed equally. EAG and JB contributed equally, had full access to all the data in the study, and take responsibility for the integrity of the data and the accuracy of the data analysis.
Corresponding authors
Ethics declarations
Competing interests
Ewan Gibb is an employee of Veracyte, Inc. The remaining authors declare no competing interests.
Ethics approval and consent to participate
Informed consent was provided by each subject and the use of the tissue was approved by the Ethics Committee of the Dana Farber Cancer Institute, Hospital del Mar and Hospital Vall d’Hebron and all research complied with local ethics guidelines.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
de Jong, J.J., Valderrama, B.P., Perera, J. et al. Non-muscle-invasive micropapillary bladder cancer has a distinct lncRNA profile associated with unfavorable prognosis. Br J Cancer 127, 313–320 (2022). https://doi.org/10.1038/s41416-022-01799-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41416-022-01799-2
This article is cited by
-
One Size Fits Some: Approaching Rare Malignancies of the Urinary Tract
Current Treatment Options in Oncology (2024)
-
Identification of exosomes-related lncRNAs in clear cell renal cell carcinoma based on Bayesian spike-and-slab lasso approach
Functional & Integrative Genomics (2023)
-
Roles of non-coding RNAs in the metabolism and pathogenesis of bladder cancer
Human Cell (2023)
-
Functions, mechanisms, and clinical applications of lncRNA LINC00857 in cancer pathogenesis
Human Cell (2023)