Abstract
Many antidepressants, atomoxetine, and several antipsychotics are metabolized by the cytochrome P450 enzymes CYP2D6 and CYP2C19, and guidelines for prescribers based on genetic variants exist. Although some laboratories offer such testing, there is no consensus regarding validated methodology for clinical genotyping of CYP2D6 and CYP2C19. The aim of this paper was to cross-validate multiple technologies for genotyping CYP2D6 and CYP2C19 against each other, and to contribute to feasibility for clinical implementation by providing an enhanced range of assay options, customizable automated translation of data into haplotypes, and a workflow algorithm. AmpliChip CYP450 and some TaqMan single nucleotide variant (SNV) and copy number variant (CNV) data in the Genome-based therapeutic drugs for depression (GENDEP) study were used to select 95 samples (out of 853) to represent as broad a range of CYP2D6 and CYP2C19 genotypes as possible. These 95 included a larger range of CYP2D6 hybrid configurations than have previously been reported using inter-technology data. Genotyping techniques employed were: further TaqMan CNV and SNV assays, xTAGv3 Luminex CYP2D6 and CYP2C19, PharmacoScan, the Ion AmpliSeq Pharmacogenomics Panel, and, for samples with CYP2D6 hybrid configurations, long-range polymerase chain reactions (L-PCRs) with Sanger sequencing and Luminex. Agena MassARRAY was also used for CYP2C19. This study has led to the development of a broader range of TaqMan SNV assays, haplotype phasing methodology with TaqMan adaptable for other technologies, a multiplex genotyping method for efficient identification of some hybrid haplotypes, a customizable automated translation of SNV and CNV data into haplotypes, and a clinical workflow algorithm.
Similar content being viewed by others
Introduction
Many antidepressants, atomoxetine, and several antipsychotics are metabolized by CYP2D6 and CYP2C19 [1,2,3,4,5,6,7]. The gene (CYP2D6) encoding the enzyme CYP2D6 is on chromosome 22q13.2 [8] adjacent to two pseudogenes, CYP2D7 and CYP2D8 [9, 10]. The high homology between CYP2D6 and these pseudogenes and the presence of flanking transposable genetic elements [11] makes the region vulnerable to the generation of variable copy numbers of the CYP2D6 gene and hybrid genes made up of sequence derived in part from CYP2D7 and in part from CYP2D6 [12,13,14,15,16,17,18,19,20,21]. Such variants are challenging to characterize for many technologies. The CYP2C19 gene encoding the CYP2C19 enzyme is located at chromosome 10q23.33, also together with other similar genes [22,23,24,25]. While structural variants of CYP2C19 have recently been identified [26], the more commonly studied haplotypes result from single nucleotide variants (SNVs) [27]. Haplotypes in both genes are referred to as “star alleles,” e.g., *2, *3, etc. as defined by PharmVar [13], a consortium which maintains a curated catalog of allelic variation in genes impacting drug metabolism, disposition, and response.
Different CYP2D6 or CYP2C19 haplotypes may be associated with different levels of enzyme activity, ranging from loss-of-function haplotypes (which give rise to no functional enzyme), to haplotypes with decreased function (which are associated with an enzyme with reduced metabolic activity), to gain-of-function haplotypes (associated with increased activity) [4]. Haplotype frequencies vary between and within ethnic groups [1, 4, 28,29,30,31]. The study of clinical associations between variants in these genes and response to relevant medications has been to date limited by the challenging nature of the genotyping, particularly in the case of CYP2D6 [32]. This gene is extremely polymorphic, with single or short sequence variants including indels (insertions/deletions), sequence derived from CYP2D7 (described as “conversions” such as an exon 9 conversion), and structural variants (deletions of the entire CYP2D6 gene, gene duplications/multiplications denoted as xNs, and hybrids [21] as above described).
There are now a number of laboratories offering testing for these genes for clinical utility; however, to date there is no consensus regarding validated methodology suitable for this purpose [33]. Clinical validation requires selecting appropriate haplotypes for testing, obtaining reference samples, and establishing test analytical validity and feasibility [34]. A recent paper provides recommendations on CYP2D6 haplotype selection for clinical testing [35], and the Genetic Testing Reference Material Program (GeT-RM) has conducted extensive work in order to provide reference samples [36,37,38]. The aims of this paper were to cross-validate multiple technologies against each other for genotyping CYP2D6 and CYP2C19 thereby facilitating feasible clinical implementation through the provision of a range of assay options, to develop customizable and automated translation of data into haplotypes, and to recommend an efficient clinical workflow algorithm that includes hybrid configurations. CYP2D6 metabolizes many other medications in addition to psychotropics (in total ~20–25% of clinically used drugs [39,40,41]); this work is also relevant for these medications, many of which are prescribed as comedications in patients with psychiatric disorders.
In the GeT-RM publications, data are available from the AmpliChip CYP450 Test, the Luminex CYP2D6 xTAG v3, and other genotyping platforms including AutoGenomics INFINITI, ParagonDx, and LDT SNaPShot, PharmacoScan, Agena MassARRAY iPLEX CYP2D6 v1.1, TaqMan assays, L-PCR, digital droplet PCR, and amplicon sequencing using next-generation sequencing (NGS) or long-read single-molecule real-time sequencing (SMRT, N = 3) [36,37,38]. CYP2D6 TaqMan assays have been compared with data arising from mPCR-RETINA, Sanger sequencing, long-PCR for CYP2D6*5, and NGS data available via the 1000 Genomes Project [42]. Validation data have been provided for the Agena VeriDose Core and CYP2D6 copy number variation (CNV) Panel versus a proprietary panel and two TaqMan assays [43]. There are two technologies that provide SMRT sequencing: Pacific Biosciences and Oxford Nanopore; the former has been compared to data for CYP2D6 from the AmpliChip CYP450 Test (which included 32 CYP2D6 variant haplotypes including some structural variants but no hybrids) on 25 individuals in one study [44], and to data from targeted Illumina NGS in 17 individuals including one hybrid haplotype (CYP2D6*36) [45]; the latter has been used on 7 reference and 25 clinical samples (which included some structural variants but no hybrids) [46]. The former was also applied to 561 patients with breast cancer, and to replication samples, although with limitations, including pertaining to hybrid haplotypes [47]. While software exists to call CYP2D6 haplotypes from next generation full sequencing data [48,49,50,51], such tools are not yet available for SMRT data, or for combinations of CNV and SNV data that arise from other technologies including NGS.
The novel contributions described herein include: (1) inter-technology concordance data on genotypes from genomic samples including a range of CYP2D6 hybrids and hybrid tandems for the AmpliChip CYP450 test, TaqMan CNV and SNV assays, the Luminex CYP2D6 and CY2C19 xTAG v3 assays, the NGS AmpliSeq Pharmacogenomics Panel, PharmacoScan, and the Agena MassARRAY (for CYP2C19); (2) details of CYP2D6 amplicon Sanger sequencing methodology including primers; (3) adaptation of the Luminex CYP2D6 assay for amplicon sequencing and provision of concordance data for this versus Sanger sequencing so that other multiplex genotyping methods can also be adapted for efficient identification of hybrid haplotypes; (4) haplotype derivation files for the interpretation of combinations of CYP2D6 CNV and SNV data including hybrids and CYP2C19; (5) development of a broader range of TaqMan SNV assays; (6) outline of haplotype phasing methodology with TaqMan adaptable for other technologies; and (7) a clinical workflow algorithm that includes hybrid configurations.
Materials and methods
Ninety-five DNA samples (originating from venous blood) were selected from those previously genotyped for CYP2D6 and CYP2C19 using the AmpliChip CYP450 Test (Roche Molecular Systems, Pleasanton, USA) supplemented by the TaqMan assay C_469857_10 for CYP2C19*17 as part of the Genome-based therapeutic drugs for depression (GENDEP) study [52]. Participants were all of self-reported White European ancestry. GENDEP was originally approved by ethics boards at all participating centers and approval for the work described herein was also provided by the University of Alberta Health Research Ethics Board—Biomedical Panel. Written informed consent was provided by all participants. The AmpliChip identified 32 CYP2D6 variant haplotypes (*2, *3, *4, *5, *6, *7, *8, *9, *10, *11, *14, *15, *17, *19, *20, *25, *26, *29, *30, *31, *35, *36, *40, *41, *114 (reported as *14A) *1×N (×N referring to more than one copy), *2×N, *4×N, *10×N, *17×N, *35×N, *41×N). In addition, it covered CYP2C19 haplotypes *2 and *3. Sample DNA concentrations were ascertained using fluorimetry-based methods (Qubit or Quantifluor).
TaqMan copy number variant (CNV) assays for CYP2D6
TaqMan CNV assays for CYP2D6 (assay IDs: Hs04083572_cn and Hs00010001_cn for intron 2 and exon 9 respectively; Thermo Fisher Scientific) were run according to the manufacturer’s protocol on a ViiA7 Real-Time PCR System (Thermo Fisher Scientific). Assays were run in quadruplicate [53]. Data were analyzed using CopyCaller software version 2.1 (Thermo Fisher Scientific) with internal calibrators of known CYP2D6 copy number according to the manufacturer’s instructions (using a confidence level of at least 95%, most being above 99%).
Samples for which the TaqMan CNV call across the two probes were not equal and hence indicative of CYP2D6 hybrids were analyzed with a third probe (assay ID Hs04502391_cn for CYP2D6 intron 6). These samples, and those representing as broad a range of CYP2D6 and CYP2C19 genotypes as possible by the AmpliChip CYP450 and TaqMan CYP2C19*17 assays, or “no call” for CYP2D6 on the AmpliChip CYP450 were then taken forward for further analysis (N = 95). The 95 were thus enriched for complex structural variants and other difficult to detect genotypes. The following genotyping techniques were employed: Luminex CYP2D6 xTAG v3 and Luminex CYP2C19 xTAG v 3, PharmacoScan (Thermo Fisher Scientific, Waltham, MA, USA), Ion AmpliSeq Pharmacogenomics Panel (Thermo Fisher Scientific), and TaqMan Drug Metabolism Genotyping Assays (Thermo Fisher Scientific). Data arising from these were then used to select samples for the generation of amplicons by long-range polymerase chain reaction (known as L-PCR) [18, 20, 54,55,56].
TaqMan SNV assays
Haplotype phasing for samples with three copies of the CYP2D6 gene according to the TaqMan, Pharmacoscan, and/or AmpliSeq CNV probe data, and heterozygous SNV data was conducted by the following methodologies: TaqMan assays for the relevant CYP2D6 SNVs on genomic DNA, and/or L-PCR specific for CYP2D6 duplicated genes followed by genotyping of the L-PCR product using relevant TaqMan SNV assays. The TaqMan SNV assays used were for CYP2D6*2, *3, *4, *6, *35 and *41 (with assay IDs C_27102425_10, C_32407232_50, C_27102431_D0, C_32407243_20, C_27102444_F0, and C_34816116_20 respectively). The TaqMan SNV assays for CYP2D6*2, *4, *10 (assay ID: C__11484460_40), and *35 were used on genomic DNA to conduct haplotype phasing for samples with CNV data consistent with three CYP2D6 genes including a hybrid gene. In addition, a TaqMan assay (assay ID C__25986767_70) was used to cross-validate a CYP2C19*2-defining SNV, rs4244285. Samples were run in duplicate on a ViiA7 Real-Time PCR System (Thermo Fisher Scientific), with genotype calling after visual inspection, outlier exclusion, and manual adjustment of CT threshold settings as necessary. Data arising from duplicates were compared with each other using an automated method available from the authors at request.
Luminex
The Luminex xTAG CYP2D6 and CYP2C19 Kits v3 (research use only versions) were run according to the manufacturer’s instructions using on a Luminex 200 system (Luminex Molecular Diagnostics, Inc., Toronto, ON, Canada). The assays use multiplex allele specific primer extension (ASPE) with a bead-based assay system. Haplotypes covered for CYP2D6 are: CYP2D6*2–*12 (including the *5 gene deletion), *14, *15, *17, *29, *35, *41, and gene duplication. Multiple different CYP2D6 haplotype translators are provided (https://doi.org/10.6084/m9.figshare.16828741) owing to the Allele Typer software having a limit on the number of haplotypes that it can process in a given translator. These cover this range of haplotypes covered by the Luminex and also permit derivation of other haplotypes of known function (*20, *39.001, *39.002, *69, *114) and sub-haplotypes (*1.011, *2.001/*2.005/*2.012/*2.013/*2.018/*2.020/*2.021, *2.004, *4.002, *4.012, *6.003, *12.001, *12.002), hybrid haplotypes (*4.013, two specific *13s (EU093102 previously known as *66, and GQ162807 previously known as *77), *36, *57, *61, *63, *68, and *83) and hybrid tandems (*4.013 + *4, *4.013×2 + *4, *4.013 + *4 × 2, *4.013 × 2 + *4 × 2, *36 + *10, *36 + *10 × 2, *36 × 2 + *10, *36 × 2 + *10 × 2). We also provide other versions of the translators in which *70 or *107 replace a *39 haplotype. These latter versions are intended for research use. The RUO software for CYP2C19 reports CYP2C19*2–*10, and *17. Our CYP2C19 haplotype translator (https://doi.org/10.6084/m9.figshare.16828738) also permits derivation of: *2.002/*2.010/*2.012, *4, *4.002, *5, *6, *7, *8, *9, and *10.
Ion AmpliSeq pharmacogenomics panel
Genotyping using the Ion AmpliSeq Pharmacogenomics Panel (Thermo Fisher Scientific) was conducted according to the manufacturer’s instructions using an Ion Chef instrument (Thermo Fisher Scientific, Waltham, MA, USA). Short stretches of genomic DNA were sequenced, including regions of CYP2D6 designed to detect CYP2D6-structural variants. Following sequencing, data were analyzed using the GeneStudio Data Analysis software (Thermo Fisher Scientific). Sequencing generated an average of 109,454 reads per sample (mean read length 142.5 bp), with two samples failing quality control (in a manner indicating likely insufficient template: mapped read numbers of 18 and 51). Variant calling by the Ion Torrent Variant Caller version 5.10.1.19 (Thermo Fisher Scientific) generated three text files: one with the genotype at each SNV (including 20 CYP2D6 variants and 11 CYP2C19 variants), one for the CYP2D6 exon 9 CNV output, and one for the CYP2D6 gene level CNV data (based on sequence across nine regions in CYP2D6) (https://doi.org/10.6084/m9.figshare.16828747). Haplotype translation files were created (using data from PharmVar, hybrid haplotype data available on the archived Human Cytochrome P450 (CYP) Allele Nomenclature Committee page [57], and a relevant publication [17]) to derive CYP2D6 and CYP2C19 haplotypes including various hybrid configurations in conjunction with the AlleleTyper software (Thermo Fisher Scientific) [58].
PharmacoScan
The PharmacoScan array-based technology was run at Neogen Genomics (Lincoln, NE, USA). The resultant data, including more than 100 variants in CYP2D6 and 60 variants in CYP2C19, were analyzed using the Axiom Analysis Suite 4.0.3.3 (Thermo Fisher Scientific). Version r8 + 20200211 of the manufacturer’s CYP2D6 haplotype translation file was used. This file was created using data from PharmVar, building on earlier work that used data available on the archived Human CYP Allele Nomenclature Committee page, and some contribution from the Aitchison laboratory. CNV calls were provided by probes for exon 9 of CYP2D6 as well as for the 5′ and 3′ flanking regions as described [38].
Long-range PCR assays with characterization of resultant amplicons
L-PCR was performed as described with minor modifications to generate an amplicon specific for the duplicated CYP2D6 gene [55]. In brief, for the L-PCR assay that generates the D amplicon (specific for duplicated CYP2D6 genes), we used primers as described [55], i.e., forward and reverse 5′-CCAGAAGGCTTTGCAGGCTTCAG-3′ and 5′-CGGCAGTGGTCAGCTAATGAC-3′, respectively, with minor modifications to the PCR conditions. Amplicons were purified by gel extraction (GeneJET Gel Extraction Kit, Thermo Fisher Scientific, Waltham, MA, USA), and genotyped using the TaqMan SNV assays described above.
Samples with unequal calls across the TaqMan, PharmacoScan, or AmpliSeq CNV probes were subjected to L-PCR assays to generate amplicons specific for CYP2D6-2D7 or CYP2D7-2D6 hybrids (E, G [20], or H [18]), with minor modifications. Amplicons were purified by gel extraction and subjected to Sanger sequencing (10 μl at 3.5 ng/μl per reaction) using BigDye Terminator version 3.1 chemistry, the Axygen CleanSEQ magnetic beads-based post-reaction clean up protocol (automated on a Biomek 3000 workstation), and a capillary 3130xl Genetic Analyzer (Thermo Fisher Scientific, Waltham, MA, USA).
Primers for sequencing (Supplementary Table 1) included novel ones designed for this study, those supplied by Dr. Gaedigk (personal communication), as well as previously reported ones used to generate the L-PCR amplicons and in prior literature [56, 59, 60]. Sequence traces were aligned (to sequences available via the PharmVar [13] or archived [57] CYP2D6 pages) and analyzed using SnapGene software version 5.1.4.1 (GSL Biotech LLC, Chicago, IL, USA).
The Agena MassARRAY
The Agena MassARRAY (Agena Bioscience, San Diego, CA, USA) uses matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry technology for resolving oligonucleotides. We ran 8 CYP2C19 variants to enable calling of 9 haplotypes. Genomic DNA was subjected to PCR followed by single-base extension with the extension products then being dispensed onto a SpectroCHIP Array and detected via mass spectrometry as described [38]. Haplotypes were assigned using Typer Analyzer software version v4.1.83 (Agena Bioscience).
Results
CYP2C19
Percentage concordance for Luminex, AmpliSeq, PharmacoScan, Agena, and prior data with consensus CYP2C19 genotype are shown in Table 1. All technologies apart from the AmpliChip were able to detect CYP2C19*6 and *8. For CYP2C19*2 and CYP2C19*17, data from all technologies, where available, were concordant.
CYP2D6
Comparative genotypic and CNV data across the technologies for samples with one and three copies of the CYP2D6 gene are shown in Supplementary Tables 2 and 3, respectively. Owing to the “no calls” in the AmpliSeq CNV data, we revised these to manual calls, where possible, after reviewing the vcf files. This did result in an improvement in the degree of concordance with consensus genotypes for the AmpliSeq. Data for samples with a CNV call of two are shown in Supplementary Table 4.
While the AmpliChip provided haplotype phasing of CYP2D6xNs (i.e., assignment of the duplication/multiplication to one or other of the two chromosomes), the other technologies included herein do not offer that. We used TaqMan assays on genomic DNA to identify which haplotype was duplicated/multiplicated based on relative magnitude of signals arising from TaqMan wild-type and mutant probes for each assay (Supplementary Fig. 1). All of our CYP2D6xNs were duplications with the exception of one sample, which had a CYP2D6*41×3. Consistent with this, our TaqMan CNV data were 4, 4, 4, and the raw PharmacoScan copy number probe calls were 4, 4, 4.
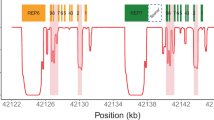
Data including consensus genotypes for the samples with hybrids genotyped to date are shown in Supplementary Table 5. Nineteen samples had an unequal call across at least two out of three CNV probesets (TaqMan, PharmacoScan, or AmpliSeq); for nine of these, the CNV pattern was consistent with a CYP2D7-2D6 hybrid, and for 10 with a CYP2D6–2D7 hybrid. For all of the CYP2D6–2D7 hybrids, the pattern was consistent with an extra CYP2D6 gene, either on the same haplotype as the hybrid gene (in cis), or on the other chromosome 22 (in trans). This was also the case for five of the CYP2D7–2D6 hybrids. Amplicons consistent with hybrids [17,18,19,20] were generated for all 19 samples. Six samples had an unequal call across CNV probes for only one platform; four of these were genotyped as CYP2D6 duplications (CYP2D6*1×2/*4, *1/*1×2, *2×2/*1 and *2×2/*35) and two as heterozygotes (*1/*2 and *1/*3.001). For three of these, amplicon G was generated; however, it should be noted that the primer pair for this amplicon will also amplify up CYP2D7 (Fig. 2 in Black et al. (2012) [20], observed where the CYP2D6 downstream gene was *1, *4, or *41; these three all had genotypes including the *1 and/or *4, specifically *1/*2, *1×2/*4, *4×2/*1).
L-PCR amplicons specific for CYP2D7–2D6 hybrids aligned well to CYP2D6*13 sequences Specifically, three samples aligned to the GQ162807 sequence (previously CYP2D6*77) deposited by Gaedigk et al. (2010) [18], where CYP2D6*13 is found in tandem arrangement with CYP2D6*2. The consensus genotype (from data including TaqMan assays) for these samples was CYP2D6*13 + *2/*1. Three other amplicons aligned well to the EU093102 sequence for another CYP2D6*13 variant, which is found as a single gene on one chromosome [15, 16], consistent with the consensus genotypes for these samples: CYP2D6*13/*4.013, CYP2D6*13/*1, and CYP2D6*13/*1 (Supplementary Table 5). At this point, we have two remaining CYP2D6*13s for which the exact CYP2D6*13 has not been identified; however, we have sufficient data to make a consensus genotype call for both (CYP2D6*13 + *4/*5, and CYP2D6*13 + *2/*41) and all CYP2D6*13 haplotypes have the same CYP2D6 enzyme activity score (zero, i.e., do not encode any functional CYP2D6 protein) owing to a T insertion in exon 1 that is a frameshift mutation resulting in premature chain termination [21]. L-PCR amplicons for CYP2D6-2D7 hybrids aligned to EU530605 (CYP2D6*4-like [17]), with one aligning to EU530606 (partial sequence for CYP2D6*68 [17]).
CYP2D6*13 + *4/*5 represents a novel haplotype. The CNV data from TaqMan and AmpliSeq were 1, 2, 2 and 1, 2 consistent with the presence of a CYP2D7–2D6 hybrid (a CYP2D6*13) with a switch region between intron 2 and intron 6. On alignment of the Sanger-sequencing data, the sequence appeared as if the sample was heterozygous from a region consistent with this inferred switch region onwards, with the region prior to this aligning well to CYP2D7. This would be consistent with our long-PCR having amplified up both a CYP2D7[REP6] (a version of CYP2D7 that has a CYP2D6 version of the repeat element at its 3′ region, consistent with the 3′ primer used for the long-PCR) and a CYP2D6*13 (legacy designations for the CYP2D6*13 haplotypes CYP2D6*67, *78, and *80 [1, 18, 19, 21, 57] have switch regions in the relevant area). Comparative data from other technologies for this sample showed genotypes of CYP2D6*4/*4 (AmpliChip), and no call with alternative calls of CYP2D6*4/UNK or *4.009/UNK (Pharmacoscan). These indicate that a CYP2D6*4 haplotype is also present. Haplotype phasing with the CYP2D6*4 TaqMan assay indicated a deletion on one allele and the CYP2D6*4 on the other. Therefore, we deduced a configuration of CYP2D6*13 + CYP2D6*4/CYP2D6*5, where the CYP2D6*5 deletion has a CYP2D7[REP6] followed by a deletion of the CYP2D6 gene.
We also subjected the L-PCR amplicons to genotyping using the Luminex CYP2D6 assay, with a protocol modification. The resultant genotypes at 14 SNVs were consistent with the sequence data (Table 2A, B). For example, the 4181G>C (rs1135840) variant was found in the Luminex and sequence data for samples aligned to GQ162807 or EU093102, while the 2851C>T (rs16947) variant was wild-type for samples aligned to EU093102 and variant for samples aligned to GQ162807. For the CYP2D6–2D7 hybrids, our CYP2D6 haplotype translator was able to identify hybrids including hybrid tandems (Supplementary Table 5).
Discussion
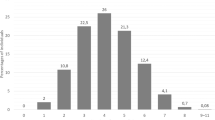
Consensus genotypes generated in 95 samples for CYP2D6 and 93 samples for CYP2C19 to date resulted in revision of assigned enzyme activity score for 28/95 (29%) and 2/93 samples (2.2%) for CYP2D6 and CYP2C19, respectively (sample selection enriched for structural variants in CYP2D6). These changes in assigned activity score were due to both changed genotype assignments and to new genotype assignments for samples that were “no calls” on AmpliChip (Fig. 1). For CYP2C19, the highest concordance with consensus genotype was in the Luminex and PharmacoScan data (100%). Data from Luminex, Agena, TaqMan, AmpliSeq, and PharmacoScan were 100% concordant for the CYP2C19*2 and CYP2C19*17, the most common loss-of-function and gain-of-function haplotypes, respectively, in individuals of European ancestry. No adjustments in the prior AmpliChip and TaqMan data were therefore required for either of these haplotypes; prior clinical association analyses conducted on the basis of these CYP2C19 haplotypes are therefore valid [52, 61].
For CYP2D6, all technologies other than the AmpliChip were able to reliably detect the CYP2D6*5. Haplotype phasing of CYP2D6xNs was achieved by using relevant TaqMan assays on genomic DNA (Supplementary Fig. 1), or by genotyping an amplicon specific for the xN. Although using allelic ratios to cluster TaqMan genotype data leaves a degree of uncertainty around genotypes (e.g., if only one probe amplifies, it may not be possible to distinguish between C/C, CC/C, CC/− and C/−), this technique can be used effectively to distinguish different heterozygote groups (Supplementary Fig. 1). A strength of the sample set was the availability of prior AmpliChip data including haplotype phasing of CYP2D6xNs. The haplotype phasing achieved with our TaqMan allelic ratio method was consistent with the prior data, where available. One sample was genotyped as having a multiplication (i.e., more copies than 2), specifically of CYP2D6*41, which has been previously described [55, 62]. The majority of the revisions in assigned enzyme activity score were due to the inability of AmpliChip to detect hybrids (Supplementary Table 5) and a sensitivity issue for CYP2D6*5 detection by AmpliChip. The latter has been previously reported [44, 53] by us and other investigators, and relates to the particular sequence used to design detection of CYP2D6*5 by the AmpliChip.
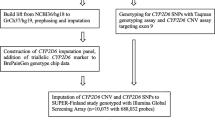
A focus of recent research on CYP2D6 is the hybrid haplotypes [17,18,19,20]. We have developed efficient methodology for characterizing a range of hybrid haplotypes: a haplotype translation tool for the interpretation of combinations of CYP2D6 CNV and SNV data including some hybrids and hybrid tandems (Supplementary Table 1), methodology for CYP2D6 Sanger sequencing, and adaptation of the Luminex CYP2D6 assay for amplicon sequencing with provision of concordance data for this versus Sanger sequencing to facilitate the application of other multiplex technologies to hybrid amplicons. In addition, we have developed a custom assay (ANT2NCE) that works on L-PCR amplicons for the sequence that occurs in CYP2D6 hybrids such as CYP2D6*36 representing a CYP2D7 exon 9 conversion. We also provide an algorithm for efficient clinical workflow that includes hybrid haplotypes including hybrid tandems (Fig. 2). The high degree of concordance between amplicon Luminex CYP2D6 and Sanger sequencing data is important and extends the coverage of the Luminex CYP2D6 assay to hybrids. Unfortunately, since we made this discovery, the assay has been withdrawn from the market. However, its components are still available for use as a laboratory developed test. Other multiplex assays currently available (such as the AmpliSeq Pharmacogenomics Panel, and the Agena MassARRAY Veridose Core plus CNV) could be likewise adapted. Of note, the workflow algorithm is capable of being adapted for CYP2C19, for which structural variants have been recently identified.
The efficient clinical workflow algorithm for CYP2D6 genotyping provided in Fig. 2 includes: multiplex SNV and CNV assays, haplotype phasing, and L-PCRs with multiplex genotyping or sequencing (Fig. 2). Appropriate positive controls (e.g., from the GeT-RM [63], especially for the haplotypes that we did not see in this European sample set and which might be found in other ethnic groups, should be run with the assays. We have provided cross-validation data between a multiplex assay and Sanger sequencing for amplicons from CYP2D6*4.013 haplotype and CYP2D6*13 haplotypes (Table 2A, B). These can act as reference data for other multiplex assays processing hybrid amplicons. Laboratories desiring to use this workflow could use one of the technologies offering multiplex SNV and CNV data (e.g., the Open Array could be supplemented by TaqMan CNV assays) on genomic DNA, with the resultant data suggesting samples with hybrid alleles; these samples would require L-PCR amplicon generation and subsequent testing using the same multiplex assay as used on the genomic DNA (adapted as necessary). Factors influencing choice of technology by a particular laboratory may include: its local availability, relevant expertize, cost, minimum and maximum number of samples per run, any required data interpretation including bioinformatics, and turnaround time. Laboratories needing to process a small number of samples rapidly (e.g., for clinicians in acute care settings) could select an assay with a low minimum number of samples per run (e.g., Luminex). Others needing to process a large number of samples rapidly could choose a large maximum number of samples per run, such as NGS or array options. The technologies have other strengths and weaknesses, e.g., in regard to their coverage of CYP2D6 CNVs. The Ion AmpliSeq Pharmacogenomics Panel provides the greatest number of regions that may be used for CYP2D6 CNV derivation; PharmacoScan provides 5′ and 3′ UTR CNV probes; and TaqMan provides CNV assays for intron 2, intron 6, and exon 9. A TaqMan CNV assay for the 5′ UTR is also available (Hs07545275_cn) [45].
To increase the breadth of CYP2D6 coverage to haplotypes in Tiers 1 and 2 of the recommendations by Pratt et al. we developed custom TaqMan assays: ANCFHM6 for rs61736512, which is part of the defining variant in CYP2D6*29, and ANWRUE2 for rs72549356 in CYP2D6*40. In regard to other haplotypes listed herein, although CYP2D6*39 is of normal function [64, 65], the enzyme activity for individuals of this haplotype in diverse ancestral populations is as yet unknown. Therefore, arguably, it can be justified to cover this haplotype in clinical genotyping. One of our samples, with a consensus CYP2D6 genotype of *10/*41 had an alternative genotype of CYP2D6*39.001/*69. The list of haplotypes covered by assays reported herein also includes haplotypes of known function but to date without reference samples (CYP2D6*12 and CYP2D6*69) [35, 66,67,68]. Should these be identified in clinical genotyping and confirmed using a second method, then reference samples could be made available to other research and clinical labs. The rationale for inclusion of CYP2D6*70 ([69]; rated as uncertain in function, with a moderate evidence level by PharmVar [13]) and CYP2D6*107 ([70]; rated as unknown function, limited evidence level [13]) is less strong. We suggest including them on a research basis to maximize potential utility gaining knowledge relevant to clinical testing in diverse populations. When we used a CYP2D6 translator with the *107 included, this resulted in several alternative calls in which a *1 was substituted by a *107.
In regard to CYP2C19, the recommendation of Pratt et al. (2018) [71] covers CYP2C19 change-of-function haplotypes of ≥1% in any ethnic group [72], which include CYP2C19*2-*10, *17, and *35). We also suggest an additional TaqMan assay (C_312628039_10) for the c.463G>T variant (rs374036992) that may be found on the CYP2C19*17 haplotype and introduces a premature stop codon [73], and an assay to enable CYP2C19*17 haplotype phasing [73].
We acknowledge several limitations of this work. Firstly, we have not covered rare variants. It has been estimated that ~6.3% of the variance in olanzapine concentration is accounted for by rare CYP2D6 variants, while rare variants are estimated to account for 4.4% of the overall genetic variability of CYP2C19 function [74]. Such variants would be identifiable by using methods that we have not validated, such as SMRT. While there are some papers reporting the use of SMRT to identify CYP2D6 variants [44, 45, 47], SMRT has however, not yet been validated on as broad a range of hybrid configurations as were included in the present report. Secondly, the work was conducted in a set of samples from European individuals being treated for depression, with samples being selected as being representative for genotypes available in the whole set and with enrichment for CYP2D6 structural variants. As such, we did not find CYP2D6 haplotypes that would be more commonly found in other ethnic groups, such as *29. Therefore although the technologies are theoretically able to identify this haplotype, our lack of detection prevented confirmation. Of note, there are reference samples for test validation available with this haplotype from the Genetic Testing Reference Material Program (GeT-RM) [63]. Thirdly, theoretically it is possible that our CNV detection methods resulted in false positive calls for copy number loss in introns 2 and 6, owing to sequence variation in the relevant regions [75]. However, as we used three different technologies (AmpliSeq, Pharmacoscan, and TaqMan), covering probes in multiple regions of CYP2D6 in addition to introns 2 and 6, and subjected any putative hybrid haplotypes to L-PCR and Sanger sequencing, we do not think this is a significant concern.
In summary, this study provides cross-validation data on a range of CYP2D6 and CYP2C19 genotypes including CYP2D6 hybrids and hybrid tandems for several assays including: AmpliChip CYP450, TaqMan CNV and SNV assays, xTAGv3 Luminex CYP2D6 and CYP2C19, the Agena CYP2C19 content from the Veridose Core, PharmacoScan, and the Ion AmpliSeq Pharmacogenomics Panel. In addition, we provide the first reference data for multiplex assay amplicon genotyping for some CYP2D6 hybrid haplotypes using long-range polymerase chain reactions (L-PCRs) followed by Sanger sequencing and Luminex. We have also developed a broader range of TaqMan SNV assays, and haplotype phasing methodology with TaqMan that is adaptable for other technologies. Finally, we have established a multiplex genotyping method for efficient identification of some hybrid haplotypes and created a customizable automated translator of SNV and CNV data for haplotype assignment. Together this work has laid the foundation for an efficient clinical workflow algorithm.
Change history
08 March 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41398-022-01845-w
References
Aitchison KJ, Jordan BD, Sharma T. The relevance of ethnic influences on pharmacogenetics to the treatment of psychosis. Drug Metab Drug Interact. 2000;16:15–38.
Lapetina DL, Yang EH, Henriques BC, Aitchison KJ. Pharmacogenomics and psychopharmacology. In: Haddad PM, Nutt DJ, editors. Seminars in clinical psychopharmacology. London: Cambridge University Press; 2020. pp. 153–204.
Jukic MM, Haslemo T, Molden E, Ingelman-Sundberg M. Impact of CYP2C19 genotype on escitalopram exposure and therapeutic failure: a Retrospective Study based on 2,087 patients. Am J Psychiatry. 2018;175:463–70.
Carvalho Henriques B, Yang EH, Lapetina D, Carr MS, Yavorskyy V, Hague J, et al. How can drug metabolism and transporter genetics inform psychotropic prescribing? Front Genet. 2020;11:491895.
Hicks JK, Swen JJ, Thorn CF, Sangkuhl K, Kharasch ED, Ellingrod VL, et al. Clinical Pharmacogenetics Implementation Consortium guideline for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants. Clin Pharmacol Ther. 2013;93:402–8.
Hicks JK, Sangkuhl K, Swen JJ, Ellingrod VL, Muller DJ, Shimoda K, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther. 2017;102:37–44.
Pharmacogenetics. Dutch Pharmacogenetics Working Group. 2019. https://www.knmp.nl/patientenzorg/medicatiebewaking/farmacogenetica/pharmacogenetics-1. Accessed 12 Sept 2021.
Gough AC, Smith CA, Howell SM, Wolf CR, Bryant SP, Spurr NK. Localization of the CYP2D gene locus to human chromosome 22q13.1 by polymerase chain reaction, in situ hybridization, and linkage analysis. Genomics. 1993;15:430–2.
Kimura S, Umeno M, Skoda RC, Meyer UA, Gonzalez FJ. The human debrisoquine 4-hydroxylase (CYP2D) locus: sequence and identification of the polymorphic CYP2D6 gene, a related gene, and a pseudogene. Am J Hum Genet. 1989;45:889–904.
Nofziger C, Turner AJ, Sangkuhl K, Whirl-Carrillo M, Agundez JAG, Black JL, et al. PharmVar GeneFocus: CYP2D6. Clin Pharm Ther. 2020;107:154–70.
Yasukochi Y, Satta Y. Evolution of the CYP2D gene cluster in humans and four non-human primates. Genes Genet Syst. 2011;86:109–16.
Gaedigk A, Ingelman-Sundberg M, Miller NA, Leeder JS, Whirl-Carrillo M, Klein TE, et al. The Pharmacogene Variation (PharmVar) Consortium: incorporation of the human cytochrome P450 (CYP) allele nomenclature database. Clin Pharmacol Ther. 2018;103:399–401.
PharmVar. CYP2D6. https://www.pharmvar.org/gene/CYP2D6. Accessed 12 Sept 2021.
Panserat S, Mura C, Gérard N, Vincent-Viry M, Galteau MM, Jacoz-Aigrain E, et al. An unequal cross-over event within the CYP2D gene cluster generates a chimeric CYP2D7/CYP2D6 gene which is associated with the poor metabolizer phenotype. Br J Clin Pharm. 1995;40:361–7.
Daly AK, Fairbrother KS, Andreassen OA, London SJ, Idle JR, Steen VM. Characterization and PCR-based detection of two different hybrid CYP2D7P/CYP2D6 alleles associated with the poor metabolizer phenotype. Pharmacogenetics. 1996;6:319–28.
Gaedigk A, Coetsee C. The CYP2D6 gene locus in South African Coloureds: unique allele distributions, novel alleles and gene arrangements. Eur J Clin Pharmacol. 2008;64:465–75.
Kramer WE, Walker DL, O’Kane DJ, Mrazek DA, Fisher PK, Dukek BA, et al. CYP2D6: novel genomic structures and alleles. Pharmacogenet Genom. 2009;19:813–22.
Gaedigk A, Fuhr U, Johnson C, Bérard LA, Bradford D, Leeder JS. CYP2D7-2D6 hybrid tandems: identification of novel CYP2D6 duplication arrangements and implications for phenotype prediction. Pharmacogenomics. 2010;11:43–53.
Gaedigk A, Jaime LK, Bertino JS Jr., Berard A, Pratt VM, Bradfordand LD, et al. Identification of novel CYP2D7–2D6 hybrids: non-functional and functional variants. Front Pharm. 2010;1:121.
Black JL 3rd, Walker DL, O’Kane DJ, Harmandayan M. Frequency of undetected CYP2D6 hybrid genes in clinical samples: impact on phenotype prediction. Drug Metab Dispos. 2012;40:111–9.
PharmVar. CYP2D6 structural variation document. 2021. https://a.storyblok.com/f/70677/x/7073c64512/cyp2d6_structural-variation_v2-1.pdf. Accessed 12 Sept 2021.
Goldstein JA, de Morais SM. Biochemistry and molecular biology of the human CYP2C subfamily. Pharmacogenetics. 1994;4:285–99.
Chen Y, Goldstein JA. The transcriptional regulation of the human CYP2C genes. Curr Drug Metab. 2009;10:567–78.
Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 2004;14:1–18.
Botton MR, Whirl-Carrillo M, Del Tredici AL, Sangkuhl K, Cavallari LH, Agúndez JAG, et al. PharmVar GeneFocus: CYP2C19. Clin Pharm Ther. 2021;109:352–66.
Botton MR, Lu X, Zhao G, Repnikova E, Seki Y, Gaedigk A, et al. Structural variation at the CYP2C locus: Characterization of deletion and duplication alleles. Hum Mutat. 2019;40:e37–e51.
PharmVar. CYP2C19. https://www.pharmvar.org/gene/CYP2C19. Accessed 12 Sept 2021.
CYP2D6 Frequency Table. Gene-specific information tables for CYP2D6 Web site. https://www.pharmgkb.org/page/cyp2d6RefMaterials. Accessed 12 Sept 2021.
CYP2D6 Allele Functionality Table. Gene-specific information tables for CYP2D6 web site. https://www.pharmgkb.org/page/cyp2d6RefMaterials. Accessed 12 Sept 2021.
CYP2C19 Frequency Table. Gene-specific information tables for CYP2C19 web site. https://www.pharmgkb.org/page/cyp2c19RefMaterials. Accessed 12 Sept 2021.
CYP2C19 Allele Functionality Table. Gene-specific information tables for CYP2C19 web site. https://www.pharmgkb.org/page/cyp2c19RefMaterials. Accessed 12 Sept 2021.
Nofziger C, Paulmichl M. Accurately genotyping CYP2D6: not for the faint of heart. Pharmacogenomics. 2018;19:999–1002.
Bousman CA, Dunlop BW. Genotype, phenotype, and medication recommendation agreement among commercial pharmacogenetic-based decision support tools. Pharmacogenom J. 2018;18:613–22.
Bousman CA, Bengesser SA, Aitchison KJ, Amare AT, Aschauer H, Baune BT, et al. Review and consensus on pharmacogenomic testing in psychiatry. Pharmacopsychiatry. 2021;54:5–17.
Pratt VM, Cavallari LH, Del Tredici AL, Gaedigk A, Hachad H, Ji Y, et al. Recommendations for clinical CYP2D6 genotyping allele selection: a Joint Consensus Recommendation of the Association for Molecular Pathology, College of American Pathologists, Dutch Pharmacogenetics Working Group of the Royal Dutch Pharmacists Association, and the European Society for Pharmacogenomics and Personalized Therapy. J Mol Diagn. 2021;23:1047–64.
Pratt VM, Zehnbauer B, Wilson JA, Baak R, Babic N, Bettinotti M, et al. Characterization of 107 genomic DNA reference materials for CYP2D6, CYP2C19, CYP2C9, VKORC1, and UGT1A1: a GeT-RM and Association for Molecular Pathology collaborative project. J Mol Diagn. 2010;12:835–46.
Pratt VM, Everts RE, Aggarwal P, Beyer BN, Broeckel U, Epstein-Baak R, et al. Characterization of 137 genomic DNA reference materials for 28 pharmacogenetic genes: a GeT–RM Collaborative Project. J Mol Diagn. 2016;18:109–23.
Gaedigk A, Turner A, Everts RE, Scott SA, Aggarwal P, Broeckel U, et al. Characterization of reference materials for genetic testing of CYP2D6 alleles: a GeT-RM Collaborative Project. J Mol Diagn. 2019;21:1034–52.
Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenom J. 2005;5:6–13.
Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharm Ther. 2013;138:103–41.
Saravanakumar A, Sadighi A, Ryu R, Akhlaghi F. Physicochemical properties, biotransformation, and transport pathways of established and newly approved medications: a systematic review of the top 200 most prescribed drugs vs. the FDA-approved drugs between 2005 and 2016. Clin Pharmacokinet. 2019;58:1281–94.
Fang H, Liu X, Ramirez J, Choudhury N, Kubo M, Im HK, et al. Establishment of CYP2D6 reference samples by multiple validated genotyping platforms. Pharmacogenom J. 2014;14:564–72.
Kothary AS, Mahendra C, Tan M, Min Tan EJ, Yi Phua JH, Gabriella G, et al. Validation of a multi-gene qPCR-based pharmacogenomics panel across major ethnic groups in Singapore and Indonesia. Pharmacogenomics. 2021;22:1041–56.
Buermans HPJ, Vossen RHAM, Anvar SY, Allard WG, Guchelaar H-J, White SJ, et al. Flexible and scalable full-length CYP2D6 long amplicon PacBio sequencing. Hum Mutat. 2017;38:310–6.
Fukunaga K, Hishinuma E, Hiratsuka M, Kato K, Okusaka T, Saito T, et al. Determination of novel CYP2D6 haplotype using the targeted sequencing followed by the long-read sequencing and the functional characterization in the Japanese population. J Hum Genet. 2021;66:139–49.
Liau Y, Maggo S, Miller AL, Pearson JF, Kennedy MA, Cree SL. Nanopore sequencing of the pharmacogene CYP2D6 allows simultaneous haplotyping and detection of duplications. Pharmacogenomics. 2019;20:1033–47.
van der Lee M, Allard WG, Vossen R, Baak-Pablo RF, Menafra R, Deiman B. et al. Toward predicting CYP2D6-mediated variable drug response from CYP2D6 gene sequencing data. Sci Transl Med. 2021;13:eabf3637.
Lee S-B, Wheeler MM, Patterson K, McGee S, Dalton R, Woodahl EL, et al. Stargazer: a software tool for calling star alleles from next-generation sequencing data using CYP2D6 as a model. Genet Med. 2019;21:361–72.
Twist GP, Gaedigk A, Miller NA, Farrow EG, Willig LK, Dinwiddie DL, et al. Constellation: a tool for rapid, automated phenotype assignment of a highly polymorphic pharmacogene, CYP2D6, from whole-genome sequences. NPJ Genom Med. 2016;1:15007.
Chen X, Shen F, Gonzaludo N, Malhotra A, Rogert C, Taft RJ, et al. Cyrius: accurate CYP2D6 genotyping using whole-genome sequencing data. Pharmacogenom J. 2021;21:251–61.
Twesigomwe D, Wright GEB, Drogemoller BI, da Rocha J, Lombard Z, Hazelhurst S. A systematic comparison of pharmacogene star allele calling bioinformatics algorithms: a focus on CYP2D6 genotyping. NPJ Genom Med. 2020;5:30.
Huezo-Diaz P, Perroud N, Spencer EP, Smith R, Sim S, Virding S, et al. CYP2C19 genotype predicts steady state escitalopram concentration in GENDEP. J Psychopharmacol. 2012;26:398–407.
Carvalho Henriques B, Buchner A, Hu X, Yavorskyy V, Wang Y, Martens K, et al. Cross-validation of technologies for genotyping CYP2D6 and CYP2C19 [Preprint]. Available from: bioRxiv https://www.biorxiv.org/content/10.1101/2019.12.24.870295v32019.12.24.870295 (2020).
Aitchison KJ, Munro J, Wright P, Smith S, Makoff AJ, Sachse C, et al. Failure to respond to treatment with typical antipsychotics is not associated with CYP2D6 ultrarapid hydroxylation. Br J Clin Pharmacol. 1999;48:388–94.
Gaedigk A, Ndjountche L, Divakaran K, Dianne Bradford L, Zineh I, Oberlander TF, et al. Cytochrome P4502D6 (CYP2D6) gene locus heterogeneity: characterization of gene duplication events. Clin Pharmacol Ther. 2007;81:242–51.
Gaedigk A, Riffel AK, Leeder JS. CYP2D6 haplotype determination using long range allele-specific amplification: resolution of a complex genotype and a discordant genotype involving the CYP2D6*59 allele. J Mol Diagn. 2015;17:740–8.
PharmVar. Archived CYP2D6 Allele Nomenclature. 2016. https://www.pharmvar.org/htdocs/archive/cyp2d6.htm. Accessed 12 Sept 2021.
CYP2D6 & CYP2C19 Haplotype Translator Tools https://www.ualberta.ca/research/innovation/license-our-technologies.html?0=cyp2d6&details=cyp2d6--cyp2c19-haplotype-translator-tools. Accessed 12 Sept 2021.
Hosono N, Kato M, Kiyotani K, Mushiroda T, Takata S, Sato H, et al. CYP2D6 genotyping for functional-gene dosage analysis by allele copy number detection. Clin Chem. 2009;55:1546–54.
Broly F, Marez D, Sabbagh N, Legrand M, Millecamps S, Lo Guidice JM, et al. An efficient strategy for detection of known and new mutations of the CYP2D6 gene using single strand conformation polymorphism analysis. Pharmacogenetics. 1995;5:373–84.
Fabbri C, Tansey KE, Perlis RH, Hauser J, Henigsberg N, Maier W, et al. Effect of cytochrome CYP2C19 metabolizing activity on antidepressant response and side effects: Meta-analysis of data from genome-wide association studies. Eur Neuropsychopharmacol. 2018;28:945–54.
Gaedigk A, Twist GP, Leeder JS. CYP2D6, SULT1A1 and UGT2B17 copy number variation: quantitative detection by multiplex PCR. Pharmacogenomics. 2012;13:91–111.
Genetic Testing Reference Materials Coordination Program (GeT-RM). Centers for Disease Control and Prevention. 2019. https://www.cdc.gov/labquality/get-rm/index.html. Accessed 12 Sept 2021.
Shimada T, Tsumura F, Yamazaki H, Guengerich FP, Inoue K. Characterization of (+/−)-bufuralol hydroxylation activities in liver microsomes of Japanese and Caucasian subjects genotyped for CYP2D6. Pharmacogenetics. 2001;11:143–56.
Hongkaew Y, Wang WY, Gaedigk R, Sukasem C, Gaedigk A. Resolving discordant CYP2D6 genotyping results in Thai subjects: platform limitations and novel haplotypes. Pharmacogenomics. 2021;22:529–41.
Marez D, Legrand M, Sabbagh N, Lo-Guidice JM, Boone P, Broly F. An additional allelic variant of the CYP2D6 gene causing impaired metabolism of sparteine. Hum Genet. 1996;97:668–70.
Gaedigk A, Twist GP, Farrow EG, Lowry JA, Soden SE, Miller NA. In vivo characterization of CYP2D6*12, *29 and *84 using dextromethorphan as a probe drug: a case report. Pharmacogenomics. 2017;18:427–31.
Gaedigk A, Frank D, Fuhr U. Identification of a novel non-functional CYP2D6 allele, CYP2D6*69, in a Caucasian poor metabolizer individual. Eur J Clin Pharm. 2009;65:97–100.
Matimba A, Del-Favero J, Van Broeckhoven C, Masimirembwa C. Novel variants of major drug-metabolising enzyme genes in diverse African populations and their predicted functional effects. Hum Genom. 2009;3:169–90.
Qiao W, Yang Y, Sebra R, Mendiratta G, Gaedigk A, Desnick RJ, et al. Long-read single molecule real-time full gene sequencing of cytochrome P450-2D6. Hum Mutat. 2016;37:315–23.
Pratt VM, Del Tredici AL, Hachad H, Ji Y, Kalman LV, Scott SA, et al. Recommendations for clinical CYP2C19 genotyping allele selection: a report of the Association for Molecular Pathology. J Mol Diagn. 2018;20:269–76.
CYP2C19. https://www.pharmgkb.org/gene/PA124/overview. Accessed 12 Sept 2021.
Skierka JM, Black JL. Analysis of compound heterozygous CYP2C19 genotypes to determine cis and trans configurations. Pharmacogenomics. 2014;15:1197–205.
Ingelman-Sundberg M, Mkrtchian S, Zhou Y, Lauschke VM. Integrating rare genetic variants into pharmacogenetic drug response predictions. Hum Genom. 2018;12:26.
Turner AJ, Aggarwal P, Boone EC, Haidar CE, Relling MV, Derezinski AD, et al. Identification of CYP2D6 haplotypes that interfere with commonly used assays for copy number variation characterization. J Mol Diagn. 2021;23:577–88.
Acknowledgements
We thank Adrian Box (Alberta Public Laboratories) for the CYP2C19 Agena data provided as a service, Kelly Homb for technical support for the Luminex assays, Lin Chen for support with interpretation of data arising from the AmpliSeq Pharmacogenomics Panel, and Carsten A. Bruckner for support for PharmacoScan data interpretation including supplying the CYP2D6 PharmacoScan allele translation file. The work reported herein was funded by: a Canada Foundation for Innovation (CFI), John R. Evans Leaders Fund (JELF) grant (32147—Pharmacogenetic translational biomarker discovery), an Alberta Innovates Strategic Research Project (SRP51_PRIME—Pharmacogenomics for the Prevention of Adverse Drug Reactions in mental health; G2018000868 to KJA and Chad Bousman), an Alberta Centennial Addiction and Mental Health Research Chair (to KJA), Alberta Innovation and Advanced Education Small Equipment Grants Program (to KJA), the Neuroscience and Mental Health Institute, Department of Psychiatry, and the Faculty of Medicine and Dentistry at the University of Alberta. Infrastructure used in this project from GP’s lab was supported by a Hotchkiss Brain Institute Dementia Equipment Fund grant (to CB and GP for the Ion) and a Canada Foundation for Innovation John R. Evans Leaders Fund Grant (CFI-JELF) (36624—Neuromuscular genetics program, to GP). GENDEP was funded by a European Commission Framework 6 grant, LSHB-CT-2003-503428. Roche Molecular Systems supplied the AmpliChip CYP450 Test arrays and some associated support. GlaxoSmithKline and the Medical Research Council (UK) contributed by funding add-on projects in the London centre. This paper represents independent research part-funded by the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care.
Author information
Authors and Affiliations
Contributions
BCH, AB, XH, YW, VY, RD, KM, MSC, BBA, JH, SS, GP, and KJA contributed to data generation and/or data analysis. BCH, AB, KW, GP, SH, and KJA were responsible for manuscript drafting, and/or revisions, and/or finalization for submission. WM, MZD, NH, JH, DS, AC, OM, MR, and KJA were responsible for patient recruitment with associated collection of DNA samples in the GENDEP study. All authors approved the final submission.
Corresponding author
Ethics declarations
Competing interests
NH has participated in research supported by CSF project No. IP-09-2014-2979. DS has received grant/research support from Janssen and Lundbeck and has served as a consultant or on advisory boards for Janssen and Lundbeck. KJA is a member of the Clinical Pharmacogenetics Implementation Consortium and the Pharmacogene Variation Consortium, has received two research grants in the last two years from Janssen Inc., Canada (fellowship grants for trainees). All other authors have nothing to disclose.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Carvalho Henriques, B., Buchner, A., Hu, X. et al. Methodology for clinical genotyping of CYP2D6 and CYP2C19. Transl Psychiatry 11, 596 (2021). https://doi.org/10.1038/s41398-021-01717-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41398-021-01717-9
This article is cited by
-
Haplotype phasing of CYP2D6: an allelic ratio method using Agena MassARRAY data
Translational Psychiatry (2024)
-
A novel AllGlo probe-quantitative PCR method for detecting single nucleotide polymorphism in CYP2C19 to evaluate the antiplatelet activity of clopidogrel
Scientific Reports (2024)
-
Current status of the analytical validation of next generation sequencing applications for pharmacogenetic profiling
Molecular Biology Reports (2023)