Abstract
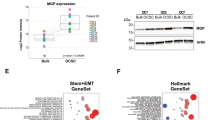
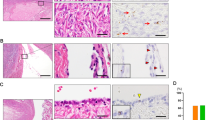
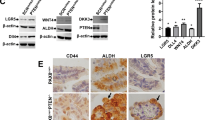
Neural precursor cell expressed, developmentally downregulated 9 (NEDD9) supports oncogenic signaling in a number of solid and hematologic tumors. Little is known about the role of NEDD9 in ovarian carcinoma (OC), but available data suggest elevated mRNA and protein expression in advanced stage high-grade cancers. We used a transgenic MISIIR-TAg mouse OC model combined with genetic ablation of Nedd9 to investigate its action in the development and progression of OC. A Nedd9−/− genotype delayed tumor growth rate, reduced incidence of ascites, and reduced expression and activation of signaling proteins including SRC, STAT3, E-cadherin, and AURKA. Cell lines established from MISIIR-TAg;Nedd9−/− and MISIIR-TAg;Nedd9+/+ mice exhibited altered migration and invasion. Growth of these cells in a syngeneic allograft model indicated that systemic Nedd9 loss in the microenvironment had little impact on tumor allograft growth, but in a Nedd9 wild-type background Nedd9−/− allografts exhibited significantly reduced growth, dissemination, and oncogenic signaling compared to Nedd9+/+ allografts. Gene expression analysis revealed that Nedd9+/+ tumors exhibited more mesenchymal “stem-like” transcriptional program, including increased expression of Aldh1a1 and Aldh1a2. Conversely, loss of Nedd9 resulted in increased expression of differentiation genes, including fallopian tube markers Foxj1, Ovgp1, and Pax8. Collectively, these data suggest that tumor cell-intrinsic Nedd9 expression promotes OC development and progression by broad induction of oncogenic protein signaling and stem/mesenchymal gene expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: A Cancer J Clin. 2016;66:7–30.
Bowtell DD, Bohm S, Ahmed AA, Aspuria PJ, Bast RC Jr., Beral V, et al. Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nat Rev Cancer. 2015;15:668–79.
McPherson A, Roth A, Laks E, Masud T, Bashashati A, Zhang AW, et al. Divergent modes of clonal spread and intraperitoneal mixing in high-grade serous ovarian cancer. Nat Genet. 2016;48:758–67.
TCGA. Integrated genomic analyses of ovarian carcinoma. Nature . 2011;474:609–15.
Bonome T, Levine DA, Shih J, Randonovich M, Pise-Masison CA, Bogomolniy F, et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68:5478–86.
Verhaak RG, Tamayo P, Yang JY, Hubbard D, Zhang H, Creighton CJ, et al. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J Clin Invest. 2013;123:517–25.
Donninger H, Bonome T, Radonovich M, Pise-Masison CA, Brady J, Shih JH, et al. Whole genome expression profiling of advance stage papillary serous ovarian cancer reveals activated pathways. Oncogene. 2004;23:8065–77.
Shagisultanova E, Gaponova AV, Gabbasov R, Nicolas E, Golemis EA. Preclinical and clinical studies of the NEDD9 scaffold protein in cancer and other diseases. Gene. 2015;567:1–11.
Wang H, Mu X, Zhou S, Zhang J, Dai J, Tang L, et al. NEDD9 overexpression is associated with the progression of and an unfavorable prognosis in epithelial ovarian cancer. Hum Pathol. 2014;45:401–8.
Seo S, Asai T, Saito T, Suzuki T, Morishita Y, Nakamoto T, et al. Crk-associated substrate lymphocyte type is required for lymphocyte trafficking and marginal zone B cell maintenance. J Immunol. 2005;175:3492–501.
Connolly DC, Bao R, Nikitin AY, Stephens KC, Poole TW, Hua X, et al. Female mice chimeric for expression of the simian virus 40 TAg under control of the MISIIR promoter develop epithelial ovarian cancer. Cancer Res. 2003;63:1389–97.
Hensley H, Quinn BA, Wolf RL, Litwin SL, Mabuchi S, Williams SJ, et al. Magnetic resonance imaging for detection and determination of tumor volume in a genetically engineered mouse model of ovarian cancer. Cancer Biol Ther. 2007;6:1717–25.
Colvin EK, Weir C, Ikin RJ, Hudson AL. SV40 TAg mouse models of cancer. Semin Cell Dev Biol. 2014;27:61–73.
Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–9.
Facciabene A, Motz GT, Coukos G. T-regulatory cells: key players in tumor immune escape and angiogenesis. Cancer Res. 2012;72:2162–71.
Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. New Engl J Med. 2003;348:203–13.
Quinn BA, Xiao F, Bickel L, Martin L, Hua X, Klein-Szanto A, et al. Development of a syngeneic mouse model of epithelial ovarian cancer. J Ovarian Res. 2010;3:24.
Gritsina G, Xiao F, O’Brien SW, Gabbasov R, Maglaty MA, Xu RH, et al. Targeted blockade of JAK/STAT3 signaling inhibits ovarian carcinoma growth. Mol Cancer Ther. 2015;14:1035–47.
Do TV, Xiao F, Bickel LE, Klein-Szanto AJ, Pathak HB, Hua X, et al. Aurora kinase A mediates epithelial ovarian cancer cell migration and adhesion. Oncogene. 2014;33:539–49.
Xu S, Yn Yang, Dong L, Qiu W, Yang L, Wang X, et al. Construction and characteristics of an E-cadherin-related three-dimensional suspension growth model of ovarian cancer. Sci Rep. 2014;4:5646.
Nikonova AS, Gaponova AV, Kudinov AE, Golemis EA. CAS proteins in health and disease: an update. IUBMB Life. 2014;66:387–95.
Izumchenko E, Singh MK, Plotnikova OV, Tikhmyanova N, Little JL, Serebriiskii IG, et al. NEDD9 promotes oncogenic signaling in mammary tumor development. Cancer Res. 2009;69:7198–206.
Singh MK, Izumchenko E, Klein-Szanto AJ, Egleston BL, Wolfson M, Golemis EA. Enhanced genetic instability and dasatinib sensitivity in mammary tumor cells lacking NEDD9. Cancer Res. 2010;70:8907–16.
Ma Y, Ma L, Guo Q, Zhang S. Expression of bone morphogenetic protein-2 and its receptors in epithelial ovarian cancer and their influence on the prognosis of ovarian cancer patients. J Exp Clin Cancer Res. 2010;29:85.
Siu MK, Wong ES, Kong DS, Chan HY, Jiang L, Wong OG, et al. Stem cell transcription factor NANOG controls cell migration and invasion via dysregulation of E-cadherin and FoxJ1 and contributes to adverse clinical outcome in ovarian cancers. Oncogene. 2013;32:3500–9.
Okada A, Ohta Y, Brody SL, Watanabe H, Krust A, Chambon P, et al. Role of foxj1 and estrogen receptor alpha in ciliated epithelial cell differentiation of the neonatal oviduct. J Mol Endocrinol. 2004;32:615–25.
Thomas J, Morle L, Soulavie F, Laurencon A, Sagnol S, Durand B. Transcriptional control of genes involved in ciliogenesis: a first step in making cilia. Biol Cell. 2010;102:499–513.
Bowen NJ, Logani S, Dickerson EB, Kapa LB, Akhtar M, Benigno BB, et al. Emerging roles for PAX8 in ovarian cancer and endosalpingeal development. Gynecol Oncol. 2007;104:331–7.
Natraj U, Bhatt P, Vanage G, Moodbidri SB. Overexpression of monkey oviductal protein: purification and characterization of recombinant protein and its antibodies. Biol Reprod. 2002;67:1897–906.
Perets R, Drapkin R. It’s totally tubular…riding the new wave of ovarian cancer research. Cancer Res. 2016;76:10–7.
Wu R, Zhai Y, Kuick R, Karnezis AN, Garcia P, Naseem A, et al. Impact of oviductal versus ovarian epithelial cell of origin on ovarian endometrioid carcinoma phenotype in the mouse. J Pathol. 2016;240:341–51.
Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507.
Schwede M, Spentzos D, Bentink S, Hofmann O, Haibe-Kains B, Harrington D, et al. Stem cell-like gene expression in ovarian cancer predicts type II subtype and prognosis. PLoS ONE. 2013;8:e57799.
Kessler M, Hoffmann K, Brinkmann V, Thieck O, Jackisch S, Toelle B, et al. The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat Commun. 2015;6:8989.
Munoz J, Stange DE, Schepers AG, van de Wetering M, Koo BK, Itzkovitz S, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012;31:3079–91.
Ice RJ, McLaughlin SL, Livengood RH, Culp MV, Eddy ER, Ivanov AV, et al. NEDD9 depletion destabilizes Aurora A kinase and heightens the efficacy of Aurora A inhibitors: implications for treatment of metastatic solid tumors. Cancer Res. 2013;73:3168–80.
Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–10.
Kim M, Gans JD, Nogueira C, Wang A, Paik JH, Feng B, et al. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269–81.
Natarajan M, Stewart JE, Golemis EA, Pugacheva EN, Alexandropoulos K, Cox BD, et al. HEF1 is a necessary and specific downstream effector of FAK that promotes the migration of glioblastoma cells. Oncogene. 2006;25:1721–32.
Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24.
Seo S, Nakamoto T, Takeshita M, Lu J, Sato T, Suzuki T, et al. Crk-associated substrate lymphocyte type regulates myeloid cell motility and suppresses the progression of leukemia induced by p210Bcr/Abl. Cancer Sci. 2011;102:2109–17.
Wang Z, Shen M, Lu P, Li X, Zhu S, Yue S. NEDD9 may regulate hepatocellular carcinoma cell metastasis by promoting epithelial-mesenchymal-transition and stemness via repressing Smad7. Oncotarget. 2016;8:1714–24.
Ratushny V, Pathak HB, Beeharry N, Tikhmyanova N, Xiao F, Li T, et al. Dual inhibition of SRC and Aurora kinases induces postmitotic attachment defects and cell death. Oncogene. 2012;31:1217–27.
Silver DL, Naora H, Liu J, Cheng W, Montell DJ. Activated signal transducer and activator of transcription (STAT) 3: localization in focal adhesions and function in ovarian cancer cell motility. Cancer Res. 2004;64:3550–8.
Ward KK, Tancioni I, Lawson C, Miller NL, Jean C, Chen XL, et al. Inhibition of focal adhesion kinase (FAK) activity prevents anchorage-independent ovarian carcinoma cell growth and tumor progression. Clin Exp Metastas. 2013;30:579–94.
Wiener JR, Windham TC, Estrella VC, Parikh NU, Thall PF, Deavers MT, et al. Activated Src protein tyrosine kinase is overexpressed in late-stage human ovarian cancers. Gynecol Oncol. 2003;88:73–9.
Sivertsen S, Berner A, Michael CW, Bedrossian C, Davidson B. Cadherin expression in ovarian carcinoma and malignant mesothelioma cell effusions. Acta Cytol. 2006;50:603–7.
Kulbe H, Chakravarty P, Leinster DA, Charles KA, Kwong J, Thompson RG, et al. A dynamic inflammatory cytokine network in the human ovarian cancer microenvironment. Cancer Res. 2012;72:66–75.
Zhong J, Baquiran JB, Bonakdar N, Lees J, Ching YW, Pugacheva E, et al. NEDD9 stabilizes focal adhesions, increases binding to the extra-cellular matrix and differentially effects 2D versus 3D cell migration. PLoS ONE. 2012;7:e35058.
Fuller E, Howell V. Culture models to define key mediators of cancer matrix remodeling. Front Oncol. 2014;4:1–7.
Buchheit CL, Weigel KJ, Schafer ZT. Cancer cell survival during detachment from the ECM: multiple barriers to tumour progression. Nat Rev Cancer. 2014;14:632–41.
Little JL, Serzhanova V, Izumchenko E, Egleston BL, Parise E, Klein-Szanto AJ, et al. A requirement for Nedd9 in luminal progenitor cells prior to mammary tumorigenesis in MMTV-HER2/ErbB2 mice. Oncogene. 2014;33:411–20.
Beck TN, Chikwem AJ, Solanki NR, Golemis EA. Bioinformatic approaches to augment study of epithelial-to-mesenchymal transition in lung cancer. Physiol Genom. 2014;46:699–724.
Paik DY, Janzen DM, Schafenacker AM, Velasco VS, Shung MS, Cheng D, et al. Stem-like epithelial cells are concentrated in the distal end of the fallopian tube: a site for injury and serous cancer initiation. Stem Cells. 2012;30:2487–97.
Connolly DC, Hensley HH. Xenograft and transgenic mouse models of epithelial ovarian cancer and non-invasive imaging modalities to monitor ovarian tumor growth in situ: applications in evaluating novel therapeutic agents. Curr Protoc Pharmacol. 2009;Chapter 14:Unit;14.2, 14.12.1–14.12.26.
Liu H, Xiao F, Serebriiskii IG, O’Brien SW, Maglaty MA, Astsaturov I, et al. Network analysis identifies an HSP90-central hub susceptible in ovarian cancer. Clin. Cancer Res. 2013;Published Online First, July 30, 2013; https://doi.org/10.1158/1078-0432.
Acknowledgements
The authors gratefully acknowledge Drs. Zeng-jie Yang and David Schlaepfer for helpful discussions; Dr. Eric Ross and Ms. Lyudmila DeMora of the FCCC Biostatistics and Bioinformatics Facility for helpful discussions and statistical analysis; Ms. Ellen Neulight for assistance with genotyping transgenic mice; Ms. Jane Miglo for technical assistance; and Mr. Xiang Hua in the Transgenic Mouse Facility for assistance with intrabursal injections. This work was supported by the FCCC Laboratory Animal, High Throughput Screening, Cell Culture, Biosample Repository, Biomedical Imaging, Flow Cytometry, Genomics, Histopathology, Transgenic Mouse and Biostatistics and Bioinformatics Facilities.
Funding
This work was supported by R01 CA136596 (to DCC); R01 CA63366 (to EAG); Ovarian SPORE P50 CA083638, the FCCC Core Grant NCI P30 CA006927, and charitable donations from the Roberta Dubrow Fund, the Teal Tea Foundation, and the Bucks County Board of Associates (to DCC).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Gabbasov, R., Xiao, F., Howe, C.G. et al. NEDD9 promotes oncogenic signaling, a stem/mesenchymal gene signature, and aggressive ovarian cancer growth in mice. Oncogene 37, 4854–4870 (2018). https://doi.org/10.1038/s41388-018-0296-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-018-0296-y
This article is cited by
-
NEDD9 links anaplastic thyroid cancer stemness to chromosomal instability through integrated centrosome asymmetry and DNA sensing regulation
Oncogene (2022)
-
NEDD9 sustains hexokinase expression to promote glycolysis
Oncogenesis (2022)
-
Rgnef promotes ovarian tumor progression and confers protection from oxidative stress
Oncogene (2019)