Abstract
The histone variant macroH2A replaces canonical H2A in the designated region of chromatin where its incorporation has the potential to establish a functionally distinct chromatin domain. The transient receptor potential canonical (TRPC) channels are a family of Ca2+-permeable cationic channels controlling changes in the cytosolic Ca2+ concentration. The proper regulation of Trpc gene expression requires chromatin remodeling, but little is known about the nature of these regulatory processes. Here, we show that macroH2A1 represses two Trpc family genes, Trpc3 and Trpc6, and attenuates Ca2+-dependent proliferative responses in bladder cancer cells. MacroH2A1 recruits histone deacetylase 1 (HDAC1) and HDAC2 to facilitate its persistent action, resulting in a compromise of histone acetylation across the Trpc3 and Trpc6 loci. Further, macroH2A1 depletion augments histone acetylation and Ca2+ influx, leading to increased cell growth and invasion. Our data provide new insights into TRPC3/TRPC6-mediated Ca2+ signaling and indicate a central role for macroH2A1 in regulating transcriptional competence of Trpc3 and Trpc6 genes.
Similar content being viewed by others
Introduction
The basic repeating unit of chromatin is the nucleosome core particle assembled from 147 base pairs of DNA wrapped around an octamer of histones H2A, H2B, H3 and H4. To properly regulate DNA-associated processes, this nuclear protein complex needs to be reorganized through various remodeling mechanisms such as histone modifications, ATP-dependent chromatin remodeling and histone variant exchange.1, 2 In contrast to canonical histones that are expressed during S phase, histone variants are expressed throughout the cell cycle and incorporated into chromatin in a replication-independent manner. MacroH2A is one of the H2A variants and is defined by the presence of a large C-terminal nonhistone domain connected to the H2A-like domain.3 There are two major macroH2A isoforms, macroH2A1 and macroH2A2, which are encoded by separate genes. Two closely-related subtypes of macroH2A1, macroH2A1.1 and macroH2A1.2, are produced by alternative splicing in the non-histone domain.4, 5 While early work focused on the role of macroH2A in the maintenance of X chromosome inactivation, more recent reports strongly implicate macroH2A in silencing of autosomal genes.6, 7, 8 To further support a broader function of macroH2A, recent biochemical studies showed that macroH2A-containing nucleosomes are refractory to ATP-dependent chromatin remodeling and histone acetylation and interfere with the binding of transcription factors to their cognate sequences.9, 10 It also has been suggested that macroH2A cooperates with negative regulators of chromatin transcription. In many of these processes, macroH2A exerts its function through physical interaction and recruitment of chromatin repressors, as illustrated by histone deacetylase (HDAC), polycomb repressive complex 2 (PRC2) and poly (ADP ribose) polymerase 1 (PARP1).6, 11, 12, 13 Of special relevance to the present study, histone acetylation is directly linked to gene activation and is dynamically governed by the opposing activities of histone acetyltransferase and HDAC. This is a potentially important point for our study, because the stable localization of HDACs at macroH2A nucleosomes helps maintain a hypoacetylated state of histones in chromatin and ultimately inactivates transcription.
Calcium (Ca2+) signaling is critical for physiological and pathological cellular responses including proliferation, migration, gene expression and contraction.14, 15 Transient receptor potential (TRP) channels constitute a large and functionally versatile family of cation-conducting channel proteins, which mediate the flux of Ca2+ across the plasma membrane and into the cytoplasm.16 The TRP canonical (TRPC) channels, a subfamily of TRP channels in mammalian cells, contain seven members, and could be divided into three groups according to sequence and functional homology: TRPC1/4/5, TRPC3/6/7 and TRPC2.17 Among these TRPC channels, TRPC3 and TRPC6 share about 70% amino-acid identity and seem to mainly contribute to cancer development and progression. For example, TRPC3/TRPC6-mediated Ca2+ signaling activates the calmodulin-dependent protein kinase and mitogen-activated protein kinase, which in turn supports the proliferative capacity of cancer cells.15 Recent studies also revealed that TRPC3 and TRPC6 channels can promote cardiac hypertrophy and that TRPC3/TRPC6 overexpression facilitates the progression of certain malignancies.18, 19, 20, 21 Given the important roles of chromatin remodeling in establishing and maintaining transcription states of genes, it is important to understand how Trpc3/Trpc6 transcription is regulated in the context of chromatin.
Here, we identify macroH2A1 as a critical regulator of TRPC3/TRPC6-mediated Ca2+ influx in bladder cancer cell lines. Transcription from Trpc3 and Trpc6 genes becomes rapidly activated when macroH2A1 is depleted. MacroH2A1 recruits HDAC1 and HDAC2, thereby abrogating histone acetylation at Trpc3 and Trpc6 genes. Supporting these results, macroH2A1 suppression reduces the recruitment of HDAC1 and HDAC2 and promotes Trpc3/Trpc6 transcription, which in turn elevates intracellular Ca2+ levels.
Results
MacroH2A1 suppresses proliferation and invasion of bladder cancer cells
As a first step toward studying cellular functions of macroH2A, we examined the expression of macroH2A1 in human bladder and prostate cell lines by western blotting. The expression levels for macroH2A1 were higher in the three bladder cell lines UROtsa, LD611 and RT4 compared with another bladder cell line J82 (Figure 1a). The prostate cell lines LNCaP and MLC also exhibited elevated levels of macroH2A1, whereas macroH2A1 expression was minimal in the two other cell lines PC3 and DU145 (Figure 1a). Since macroH2A has been proposed to suppress tumor progression via gene inactivation,22, 23, 24 we checked whether the macroH2A1 expression rate is inversely correlated with bladder cell invasiveness. The J82 cells expressing low levels of macroH2A1 exhibited more invasive potential compared with the UROtsa, LD611 and RT4 cells showing high macroH2A1 expression levels (Figure 1b).
MacroH2A inhibits bladder cancer cell proliferation and invasion. (a) Bladder cell lines (UROtsa, LD611, RT4 and J82) and prostate cell lines (MLC, LNCaP, PC3 and DU145) were lysed with RIPA buffer and subjected to western blotting. (b) The indicated bladder cell lines were detached and seeded onto the upper chamber coated with Matrigel, and then allowed to invade toward 10% FBS in the lower chamber. The graph depicts the average number of invaded cells per four fields. ND, not detected. (c, d) Proliferation of control and macroH2A1-depleted LD611 (c) and RT4 (d) cells was determined by MTT colorimetric assays. Each bar represents the mean s.d. of four replicates in three independent experiments. (e, f) Cell invasion assays performed as in (b) using control and macroH2A1-depleted LD611 (e) and RT4 (f) cells. Each bar in (b, e, f) represents the mean s.d. of three replicates in two independent experiments. **P<0.01; ***P<0.001.
To further evaluate the role of macroH2A1, we depleted macroH2A1 in the LD611and RT4 cells expressing high levels of macroH2A1 and analyzed changes in cell growth and invasiveness. In this study, it was important that macroH2A1 is depleted for prolonged periods, as this allows the study of progressive alterations of cell proliferation under identical conditions. This was achieved by using a lentiviral shRNA infection system. Western blotting and quantitative reverse transcription-PCR (qRT–PCR) confirmed that stable transfection of macroH2A1 shRNA plasmids efficiently silenced the expression of macroH2A1 in the cell (Supplementary Figure S1B). MTT assays over a period of 8 days reproducibly showed that LD611 and RT4 cells grow much more rapidly following macroH2A1 depletion (Figures 1c and d). The depletion of endogenous macroH2A1 also led to a significant increase in cell invasion compared with control cells (Figures 1e and f). These observations are consistent with the hypothesis that macroH2A1 is one of the key players governing proliferation and invasion of bladder cancer cells.
MacroH2A1 controls transcriptional programs directing Ca2+-dependent signaling pathways
Cell proliferation and invasion are complex processes that likely reflect numerous changes in multiple cellular pathways. To gain insight into the functional contributions made by macroH2A1 in the above results, we performed cDNA microarray analyses using RNA generated from control or macroH2A1-depleted LD611 cells. A comparison of the depleted and control cells indicated that 41 genes are downregulated and 169 genes are upregulated at least 1.5-fold upon macroH2A1 depletion (Supplementary Table S1). Gene ontology classification of macroH2A1 target transcripts revealed a significant enrichment in genes that are related to Ca2+ binding and endopeptidase activity (Figure 2a). To validate the microarray data, we performed qRT–PCR on 12 genes whose expression was enhanced upon macroH2A1 depletion and which are related to Ca2+ binding. As summarized in Figure 2b, there was a high correlation between the microarray data and the qRT–PCR results for all 12 genes. A noteworthy observation emerged from the microarray data was that macroH2A depletion increases the expression of Trpc3 and Trpc6 genes, but not other Trpc family genes (Figure 2c), implying that macroH2A acts as a gene-specific regulator of TRPC channels. When Ca2+ influx was compared in the depleted and control LD611 and RT4 cells, a rise in intracellular Ca2+ concentrations was evident after macroH2A1 knockdown (Figures 2d and e; Supplementary Figures S1C and D). These data strongly implicate macroH2A1 in the regulation of genes involved in Ca2+ influx. In accordance with this assertion, ectopic expression of macroH2A1.2, one of the two macroH2A1 subtypes, in LD611 cells suppressed the expression of genes encoding the 12 Ca2+ binding proteins (Figure 2i), especially TRPC3 and TRPC6, as confirmed by western blotting (Figure 2j). These results were further corroborated by Ca2+ influx assays showing that macroH2A1.2 overexpression decreased the intracellular Ca2+ concentration (Figure 2k). Furthermore, in checking whether macroH2A1 regulates the Ca2+ influx through TRPC3 and TRPC6 channels, we found that the addition of ATP, a reagent known to stimulate TRPC channels,25, 26 induced more prominent intracellular Ca2+ increase in macroH2A1-depleted cells than in control cells (Figures 2f and g). Altogether, these findings support the idea that macroH2A1 negatively affects the expression of the functional components of a Ca2+ influx pathway.
MacroH2A1 depletion enhances transcriptional potential of Ca2+ binding protein-related genes. (a) MacroH2A1-regulated genes were analyzed by DAVID bioinformatics resources (http://david.abcc.ncifcrf.gov), and ontological classification of genes based on molecular function is presented as upregulated or downregulated gene groups. (b) For validation of microarray data, 12 genes that are related to Ca2+ binding proteins and are upregulated in macroH2A1-depleted cells were subjected to qRT–PCR. Gapdh was used as an internal control gene. All expression values were normalized to the average of β-actin. (c) Trpc gene expression in control and macroH2A1-depleted LD611 cells was analyzed by qRT–PCR. ND, not detected. (d) Cell extracts from control and macroH2A1-depleted cells were immunoblotted with antibodies against TRPC3 and TRPC6. β-Actin was used as the internal control for loading. The analysis was performed in duplicates with comparable results. (e) Changes in intracellular cytosolic Ca2+ concentration after macroH2A1 depletion were measured with the Ca2+-sensitive dye Fluo-8NW. (f, g) Control and macroH2A1-delepeted LD611 cells loaded with Fura-2 AM were stimulated with 100 μM ATP. Representative traces of Ca2+ in response to ATP are shown in (f), and changes in intracellular Ca2+ were quantified in (g). (h) LD611 cells were stably transfected with control or macroH2A1.2 expression vectors, and the expression of macroH2A1.2 at the mRNA and protein levels was analyzed by qRT–PCR (left) and western blotting (right). (i) qRT–PCR was performed to check relative expressions of Ca2+ binding-related genes, which are downregulated after macroH2A1.2 expression. (j) TRPC3 and TRPC6 protein levels in control and macroH2A1.2-transfected cell were evaluated by western blotting. (k) The intracellular Ca2+ concentration was determined as in (e), but after macroH2A1.2 expression. Each bar in (b, c, e, g–i, k) represents the mean s.d. of three replicates in two independent experiments. *P<0.05; **P<0.01;***P<0.001.
MacroH2A1 and TRPC3/TRPC6 are reciprocally expressed in bladder cancer specimens
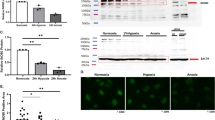
Our microarray results suggested that macroH2A1 negatively regulates genes related to Ca2+ binding proteins. Among these genes, Trpc3 and Trpc6 are of particular interest due to their critical roles in regulating extracellular Ca2+ influx. To investigate whether macroH2A1 expression inversely correlates with TRPC3/TRPC6 expression in bladder cancer, we analyzed their expression patterns in tissue microarrays containing 36 bladder cancer specimens and 12 normal tissues. Representative results of immunohistochemical staining for macroH2A1, TRPC3 and TRPC6 are shown in Figure 3a. The staining for macroH2A1 in the majority of grade 3 bladder tumor tissues (∼83%) was weak or negative, while the staining was moderate or strong in most normal (∼66%) and low-grade (grades 1 and 2) tumor tissues (∼57%) (Figure 3b, left; Supplementary Table S2). On the contrary, moderate to strong staining of TRPC3 was detected in about 90% of bladder tumor tissues, but only in circa 50% of normal tissues (Figure 3b, middle; Supplementary Table S2). Moderate to strong staining intensity for TRPC6 was also evident in high-grade (grade 3) tumor tissues (Figure 3b, right; Supplementary Table S2). Consistent with these results, our western blot analyses of three bladder (LD611, RT4 and J82) and two prostate cancer cell lines (PC3 and DU145) showed higher expression of TRPC3 and TRPC6 compared with their normal counterparts (Supplementary Figure S1E). Overall, these results are in good agreement with the gene expression data (Figure 2) and suggest that low macroH2A1 levels coincide with elevated TRPC3/TRPC6 expression in a large group of bladder tumor specimens.
Immunohistochemical staining of macroH2A1, TRPC3 and TRPC6 in tissue microarray. (a) Tissue microarrays containing 36 cases of bladder tumor with 12 normal tissues were subjected to immunohistochemistry with antibodies against macroH2A1, TRPC3 and TRPC6. High-power magnifications are shown for representative immunostaining samples. Bar, 50 μm. (b) Immunostaining scores of macroH2A1, TRPC3 and TRPC6 in bladder normal and malignant tissues. The graph indicates the percentage of sections with different scores (negative, weak, moderate and strong).
MacroH2A1 impairs Trpc3/Trpc6 transcription by facilitating HDAC1/HDAC2 recruitment and inhibiting histone acetylation
Given demonstrated suppression of Trpc3 and Trpc6 genes by macroH2A1, we next investigated the distribution of macroH2A1 across the Trpc3 and Trpc6 loci by chromatin immunoprecipitation (ChIP) assays. Cross-linked chromatin was isolated from control and macroH2A1-depleted cells, and macroH2A1 occupancy was probed for two upstream regions (regions A and B), transcription start site (TSS; region C) and two coding regions (regions D and E) (Figure 4a; Supplementary Figure S2A). We detected high macroH2A1 signals upstream (regions A and B) and downstream (regions D and E) of the TSS, but reduced signals at TSS (region C), in both the Trpc3 and Trpc6 loci (Figure 4b; Supplementary Figure S2B). These observations are in agreement with data from recent genome-wide studies that demonstrated the minimal presence of macroH2A in the vicinity of TSSs.6
MacroH2A1-dependent interplay between HDAC1 and HDAC2 in suppressing Trpc6 gene. (a) Approximate locations of five amplicons at the Trpc6 locus used in the ChIP assays are shown. (b–e) LD611 cells were depleted of macroH2A1, and ChIP experiments were performed using antibodies against macroH2A1 (b), H3ac (c), HDAC1 (d) and HDAC2 (e). Precipitation efficiencies relative to non-enriched input samples were determined for the five locations across the Trpc6 locus by qPCR with primers depicted in (a) and listed in Supplementary Table S3. (f) qRT–PCR and western blot analyses demonstrate the level of TRPC6 in LD611 cells expressing control shRNA and shRNAs targeting HDAC1 and HDAC2. (g–i) ChIP assays using an antibody recognizing H3ac were performed on samples from control cells and cells depleted of HDAC1 and/or HDAC2. (j, k) LD611 cells depleted of HDAC1 or HDAC2 were subjected to ChIP assays of Trpc6 gene using antibodies against HDAC2 (j) and HDAC1 (k). Each bar represents the mean s.d. of three replicates in two independent experiments. *P<0.05; **P<0.01; ***P<0.001.
When ChIP experiments were performed on macroH2A1-depleted cells, almost complete loss of macroH2A1 was detected at the Trpc3 and Trpc6 loci (Figure 4b; Supplementary Figure S2B). Because macroH2A is known to interfere with histone acetylation,10, 11 the levels of H3 acetylation were also determined. Expectedly, macroH2A1 depletion resulted in a distinct increase in H3 acetylation around the TSS at the Trpc6 locus (Figure 4c). Interestingly, however, H3 acetylation was increased across the Trpc3 locus in response to macroH2A1 depletion (Supplementary Figure S2C). These data raised the question of which HDACs are responsible for the observed changes in histone acetylation. It has been shown that HDAC1 and HDAC2 can directly interact with the nonhistone region of free macroH2A.11 Thus, macroH2A may contribute to the recruitment of HDAC1/HDAC2 and alter the state of histone acetylation in chromatin. In fact, the predicted roles of macroH2A are supported by the observation that HDAC1/HDAC2 occupancy at the Trpc3 and Trpc6 loci is significantly attenuated after macroH2A1 depletion (Figures 4d and e; Supplementary Figures S2D and E). ChIP experiments on the Trpc1 and Trpc4 genes whose expression was not affected upon macroH2A1 depletion detected very low levels of macroH2A1, HDAC1 and HDAC2 in all reactions (Supplementary Figures S3A–C).
The finding that both HDAC1 and HDAC2 occupy at the Trpc3 and Trpc6 loci prompted us to check whether they are functionally linked to macroH2A-induced suppression of the genes. To this end, LD611 cells were transfected with the lentiviral vectors encoding HDAC1 and HDAC2 shRNAs individually or simultaneously (Supplementary Figure S3D), and then the effects on TRPC3 and TRPC6 expression were measured. From our qRT–PCR and western blot analyses, no observable changes in TRPC3 and TRPC6 expression were detected after single knockdown of HDAC1 or HDAC2 (Figure 4f; Supplementary Figure S2F). Remarkably, however, much higher levels of TRPC3 and TRPC6 expression were detected when both HDAC1 and HDAC2 were depleted (Figure 4f; Supplementary Figure S2F), indicating compensatory mechanisms between the two HDACs in repressing Trpc3 and Trpc6 genes.
To generate further insight into the function of HDAC1 and HDAC2, we determined whether HDAC1 and HDAC2 are responsible for macroH2A1-induced histone deacetylation at the Trpc3 and Trpc6 loci. When ChIP experiments were performed using cells depleted of HDAC1 or HDAC2, no measurable effect on H3 acetylation was detected (Figures 4g and h; Supplementary Figures S2G and H). However, depletion of both HDAC1 and HDAC2 elevated H3 acetylation levels at the Trpc3 and Trpc6 loci (Figure 4i; Supplementary Figure S2I), again reflecting compensatory functions of HDAC1 and HDAC2 in perturbing histone acetylation at these two loci. In accordance with this idea, HDAC1 depletion from Trpc3 and Trpc6 genes by RNAi coincided with a distinct increase in HDAC2 localization, whereas HDAC2 depletion did so for HDAC1 localization, at the same loci (Figures 4j and k; Supplementary Figures S2J, K, S3E and F).
HDAC1 and HDAC2 preferentially interact with macroH2A1 nucleosomes
On the basis of the fact that macroH2A1 depletion attenuates the recruitment of HDAC1/HDAC2 to the Trpc3 and Trpc6 loci, we postulated that HDAC1/HDAC2 can bind macroH2A1 nucleosomes. To check this possibility, LD611 cells were transfected with expression vectors for Flag-macroH2A1.2 and Flag-H2A. After confirming the equivalent expression of the constructs, soluble chromatin fragments were prepared from cell nuclei and digested with micrococcal nuclease (MNase) to yield mainly mononucleosomes. Mononucleosomes containing ectopic macroH2A1.2 or H2A were immunoprecipitated with anti-Flag antibody. Coomassie blue staining confirmed that purified nucleosomes retain about equal amounts of ectopic macroH2A1.2 and H2A (Figure 5a). Western blotting of isolated nucleosomes revealed that both HDAC1 and HDAC2 bind macroH2A1.2 nucleosomes and that their binding decreases the level of H3 acetylation (Figure 5b). Because there was no detectable binding of HDAC1 and HDAC2 to H2A nucleosomes, the observed binding of HDAC1 and HDAC2 was specific. Additionally, the endogenous-free macroH2A1 displayed HDAC1/HDAC2-binding properties that were similar to those of macroH2A1 nucleosomes (Figure 5c), suggesting that HDAC1 and HDAC2 interact with macroH2A1 nucloesomes through macroH2A1.
HDAC1 and HDAC2 bind to the nonhistone domain of macroH2A1. (a) LD611 cells were transfected with control, H2A and macroH2A1.2 expression vectors, and mononucleosomes were prepared by MNase digestion as recently described.35 Mononucleosomes containing ectopic histones were purified by anti-Flag immunoprecipitation. Histone compositions of the purified nucleosomes were analyzed by 15% SDS–PAGE with Coomassie brilliant blue staining. (b) LD611 cells transfected with the indicated expression vectors were subjected to pull down with anti-Flag M2 agarose, and the bound proteins were detected by western blotting. (c) Whole cell extracts were prepared from LD611 cells and immunoprecipitated with anti-macroH2A1 antibody. The precipitates were analyzed by western blotting with antibodies against HDAC1, HDAC2 and macroH2A1 as indicated. (d) MacroH2A1.2 was incubated with GST-full length HDAC1 or GST-HDAC1 deletion mutants immobilized on glutathione Sepharose 4B. After extensive washing, the presence of macroH2A1.2 in the beads was determined by western blotting with anti-macroH2A1 antibody. Input corresponds to 5% of the macroH2A1.2 used in the binding reactions. Numbers indicate amino-acid residues. (e) GST pull-down assays were carried out as in (d), but with GST-fused full-length HDAC2 or its deletion mutants immobilized on glutathione Sepharose beads. (f) HDAC1 and HDAC2 were incubated with GST-full length macroH2A1.2 or GST-macroH2A1.2 deletion mutants. Binding of HDAC1 and HDAC2 was analyzed by western blotting. Lanes 1 and 6 represent 5% of the input. Asterisks in (d–f) indicate non-specific bands.
To gain support for the binding results described above, we conducted in vitro pull-down assays using bacterially produced macroH2A1.2, HDAC1 and HDAC2. As shown in Figures 5d and e, both GST-HDAC1 and GST-HDAC2 interacted with His-macroH2A1.2, whereas GST alone did not. In similar binding experiments with three distinct regions of HDAC1 and HDAC2, the C-terminal regions, but not the N-terminal regions and catalytic domains, efficiently interacted with macroH2A1.2 (Figures 5d and e). Additionally, in mapping HDAC1/HDAC2-interacting region of macroH2A1.2, the binding of the non-histone domain (residues 123–371) was readily detectable, but the N-terminal H2A-like domain (residues 1–122) showed no apparent interaction with HDAC1/HDAC2 (Figure 5f). Moreover, macroH2A1.2 fragment containing residues 123–180 bound HDAC1/HDAC2 as efficiently as the full-length protein, reinforcing conclusion that the primary HDAC1/HDAC2-binding capacity of macroH2A1.2 resides in this region.
MacroH2A1 drives the observed cellular changes in TRPC3/TRPC6-dependent manner
Several findings suggest that macroH2A inhibits cell growth and invasion by targeting Trpc3 and Trpc6 genes that control Ca2+ influx. First, among the genes that are implicated in the Ca2+ influx pathway, Trpc3 and Trpc6 genes are selectively upregulated in macroH2A1-depleted cells. Second, the ChIP signal for H3 acetylation, a hallmark of active transcription, is enhanced at Trpc3 and Trpc6 genes after macroH2A1 depletion. Third, macroH2A1 is necessary for the recruitment of HDAC1/HDAC2 to Trpc3 and Trpc6 genes. Fourth, macroH2A1 depletion stimulates Ca2+ influx and increases the intracellular Ca2+ concentration. To determine whether the observed effects of macroH2A1 depend on TRPC3 and TRPC6, we depleted them in LD611 cells (Supplementary Figures S4A, B and S5A) and measured cell growth and invasion. As summarized in Figure 6a, TRPC6 depletion decreased cell viability considerably, and TRPC3 depletion did so moderately. Simultaneous depletion of TRPC3 and TRPC6 produced more prominent reduction in cell viability (Supplementary Figure S5B). The observed effects of TRPC3/TRPC6 silencing largely disappeared upon concomitant depletion of macroH2A1, confirming the functional link between macroH2A1 and TRPC3/TRPC6. Consistent with the results of cell viability assays, LD611 cell invasion was also reduced severely after individual or simultaneous knockdown of TRPC3 and TRPC6 (Figure 6b; Supplementary Figure S5C). Simultaneous suppression of macroH2A1 reduced the effects of TRPC3/TRPC6 depletion on LD611 cell invasion. On the basis of these observations, we favor a model whereby macroH2A1 inhibits cell growth and invasion through selectively perturbing the expression of Trpc3 and Trpc6 rather than other genes.
TRPC3/TRPC6 silencing results in loss of macroH2A1 function. (a) Cell proliferation assays were carried out in quadruplicate using cells depleted of TRPC3, TRPC6 and/or macroH2A1 as indicated. Each bar represents the mean s.d. of four replicates in three independent experiments. (b) Cell invasion assays were performed using cells depleted of TRPC3, TRPC6 and/or macroH2A1. Each bar represents the mean s.d. of three replicates in two independent experiments. *P<0.05; **P<0.01; ***P<0.001. (c) Model of Trpc3/Trpc6 gene regulation by macroH2A and HDAC1/HDAC2. In response to repressive cellular signals, macroH2A is incorporated into the Trpc3 and Trpc6 loci. The non-histone domain of macroH2A physically interacts with HDAC1 and HDAC2, and this interaction is proposed to recruit HDAC1 and HDAC2 to the Trpc3 and Trpc6 loci. This leads to histone deacetylation and Trpc3/Trpc6 gene silencing. See DISCUSSION for more details.
Discussion
Although macroH2A-induced gene silencing has been implicated in the regulation of cancer initiation and progression, precise mechanisms connecting macroH2A to these processes are only beginning to emerge. In the current study, we used human bladder cancer cells and identified macroH2A1 as a key transcriptional repressor of Trpc3 and Trpc6 genes, whose expression is crucial for establishing a Ca2+-permeable pathway in the plasma membrane. MacroH2A1 not only interferes with the spreading of histone acetylation over Trpc3 and Trpc6 genes, but also facilitates the recruitment of HDAC1/HDAC2 at the genes (see Figure 6c). To our knowledge, this is the first study to provide an example of chromatin remodeling processes regulating expression of ion channel genes and to show the functional cooperativity between macroH2A1 and HDAC1/HDAC2.
To investigate the role of macroH2A1 as a specific transcriptional modulator, we compared gene expression profiles of macroH2A1-depleted bladder cancer cells with that of mock-depleted cells. There were many changes in the gene expression profile of bladder cancer cells after macroH2A depletion, and the molecular functions of the altered genes were especially enriched for functions that contribute to Ca2+-dependent cell proliferative responses. Among most significantly altered genes, we selected Trpc3 and Trpc6 for further study because their gene products are most directly associated with Ca2+ entry pathway. In probing five regions in Trpc3 and Trpc6 genes by ChIP assays, we detected macroH2A1 occupancy both upstream and downstream of the genes. These findings are in keeping with previous studies indicating that macroH2A is widely distributed over the genome and regulates transcriptional competence of chromatin in a large scale.6, 7, 27
In further support of a repressive role for macroH2A, coincidental appearance of H3 acetylation was detected across the Trpc3 locus under macroH2A1-depleted conditions. Notably, however, TRPC6 transactivation observed in macroH2A1-depleted cells was accompanied by H3 acetylation in the vicinity of TSSs. We speculate that the acetylation signals around the 5′ ends of the gene are sufficient to prevent the repressive effects of macroH2A1, which is distributed throughout the gene. On the basis of our results and the results from previous studies,10, 11 macroH2A1-mediated gene silencing we observed appears to be at least partly through protection of histone tails against histone acetyltransferase activities by the non-histone domain of macroH2A1. Although a positive signal in ChIP analysis using macroH2A1-depleted cells strongly indicates the presence of histone acetylation, one can argue that the lack of the acetylation signal in control cells may reflect a low accessibility of histone tails in macroH2A1 nucleosomes rather than the absence of the modification. However, the fact that macroH2A1 depletion minimally alters histone methylations at H3K4, H3K9 and H3K27 in our ChIP experiments constitutes a powerful argument against this possibility (Supplementary Figures S6A–C).
Another important finding of this study is that macroH2A1 cooperates with HDAC1 and HDAC2 to maintain Trpc3 and Trpc6 genes in an inactive state. This finding is in line with previous in vitro data showing the direct interaction between macroH2A and HDAC1/HDAC2.11 Additionally, our functional studies showed an apparent redundancy of HDAC1 and HDAC2, and established that they have compensatory functions in macroH2A1-mediated repression of Trpc3 and Trpc6 genes. This finding is consistent with there being a tight relationship between macroH2A exchange and repressive histone modifications.28 While it is generally recognized that HDACs can affect chromatin transcription through changes in the rate of histone acetylation, we cannot conclusively rule out an indirect effect of HDACs on gene transcription. However, the finding that HDAC1 and HDAC2 selectively associate with unacetylated macroH2A1 nucleosomes strongly suggests that chromatin repression generated by HDAC1 and HDAC2 is dependent on histone deacetylation in their target sites.
Recently, Kapoor et al.22 reported that transcriptional repression of CDK8, a subunit of the mediator subcomplex, is a critical component of macroH2A-dependent suppression of melanoma cell proliferation. Thus, it might be expected that CDK8 would be a potential target for macroH2A1 in bladder cancer cells and would be involved in macroH2A1-mediated growth suppression. However, in examining this possibility, we did not observe any effects of macroH2A1 depletion on CDK8 expression. MacroH2A1 expression and its location across the genome are likely to be different in the melanoma cells used in the CDK8 study and the LD611 bladder cells used in this study. Also, there is a varied expression and localization of HDAC1/HDAC2 in different cancer cells that could have dramatic effects on the ability of macroH2A to function on a range of targets. Changes in the cellular level of macroH2A were also detected in lung cancer and implicated in cell proliferation partly through lowering PARP1 protein levels.23 These data suggest that macroH2A1-mediated suppression of cancer cell growth employs multiple mechanisms and targets depending on cell types, which will need to be addressed in more detail in future studies.
Previous studies have shown that TRPC3 and TRPC6 channels contribute to growth and proliferation of different types of cancer cells, like prostate, breast, liver and brain.18, 19, 20, 21, 29, 30 The current study further advance these previous findings on bladder cancer cells, and indicates that TRPC3 and TRPC6 are key regulators of Ca2+-mediated cell proliferation. In fact, the findings described in this study provide the first demonstration that macroH2A1 exchange and HDAC1/HDAC2-mediated histone deacetylation at Trpc3 and Trpc6 genes contribute to the regulation of Ca2+ influx. Nonetheless, many interesting questions remain to be answered. For instance, it is unknown whether other chromatin remodeling activities, in addition to the identified interplay between macroH2A and HDAC1/HDAC2, are involved in the formation of constitutively repressed states of Trpc3 and Trpc6 genes or other Trpc members. Because histone acetyltransferase activates chromatin transcription, opposite to the roles of macroH2A and HDAC1/HDAC2, future work examining the regulation of Trpc genes in Ca2+ influx should also focus on the competitive action of histone acetyltransferases against macroH2A and HDAC1/HDAC2.
Materials and methods
Cell lines, expression vectors and antibodies
LNCaP and PC3 cells were maintained routinely in RPMI culture media with 10% heat-inactivated fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA). LD611, DU145, J82 and UROtsa cells were grown in DMEM containing 10% FBS (LD611, DU145 and J82 cells) or 5% FBS (UROtsa cells). RT4 and MLC cells were cultured in 10% FBS-supplemented McCOY’s 5A and 10% FBS-supplemented T medium, respectively. For mammalian expression of H2A and macroH2A1.2, the corresponding cDNAs were ligated into the correct reading frame of pIRES containing Flag coding sequence. Bacterial expression vectors encoding macroH2A1.1, macroH2A1.2, macroH2A2, HDAC1 and HDAC2 were generated by PCR tagging of the corresponding cDNA and subcloning them into pGEX-4T1 or pET-11d vector in frame with 5′ GST or hexa-His sequences. Further details of plasmid constructions are available upon request. Affinity-purified rabbit macroH2A1 antibody has been described previously,31 and as reconfirmed in our study (Supplementary Figure S1A), this antibody recognizes both macroH2A1.1 and macroH2A1.2, but not macroH2A2. Commercial antibodies used in this study are as follows: TRPC3 and TRPC6 antibodies from Alomone labs (Jerusalem, Israel), macroH2A2 and H3 antibodies from Abcam (Cambridge, MA, USA), acetylated H3 (H3ac) antibody from Millipore (Billerica, MA, USA), Actin antibody from Sigma (St Louis, MO, USA), His tag and GST antibodies from Santa Cruz (Dallas, TX, USA) and HDAC1 and HDAC2 antibodies from Active Motif (Carlsbad, CA, USA).
Co-immunoprecipitation
For co-immunoprecipitation of endogenous macroH2A1 and HDAC1/HDAC2, LD611 cells were washed with cold phosphate-buffered saline and lysed in buffer containing 50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 mM PMSF and 1 × protease inhibitor cocktail (Roche, Mannheim, Germany). Lysates (2 mg) were incubated with macroH2A1 antibody overnight on ice followed by incubation with Protein A/G agarose beads for 90 min. Beads were washed four times with lysis buffer, and the binding of HDAC1/HDAC2 was analyzed by western blotting.
RNA interference
DNA oligonucleotides encoding shRNAs specific for macroH2A1, HDAC1, HDAC2, TRPC3 and TRPC6 were annealed and ligated into the lentiviral mammalian vector pLKO.1 (Addgene, Cambridge, MA, USA) according to standard procedures. Cells were transfected with shRNA constructs against macroH2A1 sh1 (5′-CTGAACCTTATTCACAGTGAA-3′), macroH2A1 sh2 (5′-GCCAATGATGAAGAGCTGAAT-3′), HDAC1 (5′-CGTTCTTAACTTTGAACCATA-3′), HDAC2 (5′-CAGTCTCACCAATTTCAGAAA-3′), TRPC3 (5′-CCCAGTTTACATGGACTGAAA-3′), TRPC6 (5′-CCTGGGTAATAGGCATGATAT-3′) or control (5′-CAACAAGATGAAGAGCACCAA-3′) along with packaging plasmid (pCD/NL-BH) and envelope plasmid (pCMV/VSV-G) using the Lipofectamine reagent (Invitrogen) for 2 days. Cells (1 × 105) were infected with virus particles containing macroH2A1, HDAC1, HDAC2, TRPC3, TRPC6 or control shRNAs and selected with puromycin (2 μg/ml) for 2 weeks.
Tissue microarrays and immunohistochemistry
Bladder tissue microarrays, containing 36 cases of bladder malignant tissues and 12 cases of normal tissues, were purchased from U.S. Biomax Inc. (Rockville, MD, USA). The formalin-fixed, paraffin-embedded sections were stained for macroH2A1.2, TRPC3 and TRPC6 following standard immunohistochemistry protocols, as described.19 The staining intensity was divided into four categories: negative or marginal staining (<20% of cells), weak staining (20–50% of cells), moderate staining (50–80% of cells) and strong staining (>80% of cells).
Microarray, qRT–PCR and ChIP assays
Two independent RNA samples were prepared from control and macroH2A-depleted LD611 cells using the TRIzol reagent (Invitrogen), and transcript analysis was carried out using a whole-genome expression array (Sentrix Human-6 Expression BeadChip version 3, Illumina, Hayward, CA, USA) as recently described.32 Differential gene expression and statistical analyses were carried out using the software ArrayPipe (www.pathogenomics.ca/arraypipe) as previously described.33 The qRT–PCR was performed using the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA) and the IQ SYBR Green Supermix (Bio-Rad) with an iCycler IQ5 real time cycler (Bio-Rad). ChIP assays were performed in LD611 cells using the ChIP assay kit (Millipore) as recently described.34 The primers used for qRT–PCR and ChIP assays are listed in Supplementary Table S3.
Cell proliferation and invasion assays
Cell proliferation was measured by MTT assay as reported.35 For cell invasion assay, cells were harvested and suspended in culture medium containing 5% FBS and then seeded to the upper chamber coated with Matrigel (BD Biosciences, San Jose, CA, USA). Cells were allowed to invade toward 10% FBS in the lower chamber for 48 h. The invaded cells on the underside of the transwell filters were fixed with 10% formaldehyde for 15 min and stained with 1% crystal violet for 1 h. Cells were photographed and counted.
Ca2+ influx assay
For assays with the Fluo-8 NW dye, control and macroH2A1-depleted cells were cultured separately in a 96-well plate. The growth medium was replaced with 100 μl/well Fluo-8 dye solution containing probenecid to prevent extrusion of the dye out of cells. The plates were maintained at 37 °C for 30 min and then at room temperature for an additional 30 min. The changes in Fluo-8 NW fluorescence were measured using a Plate Chameleon V plate reader (Turku, Finland) with excitation at 490 nm, and emission measured at 525 nm. For assays with the Fura-2 AM, cells were seeded on coverslips and incubated with 2 μM Fura-2 AM at 37 °C for 50 min in Hepes-buffered solution. Cells were imaged using a F4500 fluorescence spectrophotometer (Hitachi, Tokyo, Japan) with dual excitation wavelengths of 340 and 380 nm and detection of fluorescent emission at 510 nm.26
References
Talbert PB, Henikoff S . Histone variants—ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol 2010; 11: 264–275.
Wang GG, Allis CD, Chi P . Chromatin remodeling and cancer, Part II: ATP-dependent chromatin remodeling. Trends Mol Med 2007; 13: 373–380.
Pehrson JR, Fried VA . MacroH2A, a core histone containing a large nonhistone region. Science 1992; 257: 1398–1400.
Chadwick BP, Willard HF . Histone H2A variants and the inactive X chromosome: identification of a second macroH2A variant. Hum Mol Genet 2001; 10: 1101–1113.
Costanzi C, Pehrson JR . MACROH2A2, a new member of the MARCOH2A core histone family. J Biol Chem 2001; 276: 21776–21784.
Buschbeck M, Uribesalgo I, Wibowo I, Rue P, Martin D, Gutierrez A et al. The histone variant macroH2A is an epigenetic regulator of key developmental genes. Nat Struct Mol Biol 2009; 16: 1074–1079.
Changolkar LN, Singh G, Cui K, Berletch JB, Zhao K, Disteche CM et al. Genome-wide distribution of macroH2A1 histone variants in mouse liver chromatin. Mol Cell Biol 2010; 30: 5473–5483.
Costanzi C, Pehrson JR . Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 1998; 393: 599–601.
Angelov D, Molla A, Perche PY, Hans F, Cote J, Khochbin S et al. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol Cell 2003; 11: 1033–1041.
Doyen CM, An W, Angelov D, Bondarenko V, Mietton F, Studitsky VM et al. Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol Cell Biol 2006; 26: 1156–1164.
Chakravarthy S, Gundimella SK, Caron C, Perche PY, Pehrson JR, Khochbin S et al. Structural characterization of the histone variant macroH2A. Mol Cell Biol 2005; 25: 7616–7624.
Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol 2009; 16: 923–929.
Nusinow DA, Hernandez-Munoz I, Fazzio TG, Shah GM, Kraus WL, Panning B . Poly(ADP-ribose) polymerase 1 is inhibited by a histone H2A variant, MacroH2A, and contributes to silencing of the inactive X chromosome. J Biol Chem 2007; 282: 12851–12859.
Clapham DE . Calcium signaling. Cell 2007; 131: 1047–1058.
Kahl CR, Means AR . Regulation of cell cycle progression by calcium/calmodulin-dependent pathways. Endocr Rev 2003; 24: 719–736.
Clapham DE, Runnels LW, Strubing C . The TRP ion channel family. Nat Rev Neurosci 2001; 2: 387–396.
Vazquez G, Wedel BJ, Aziz O, Trebak M, Putney JW Jr. . The mammalian TRPC cation channels. Biochim Biophys Acta 2004; 1742: 21–36.
Chigurupati S, Venkataraman R, Barrera D, Naganathan A, Madan M, Paul L et al. Receptor channel TRPC6 is a key mediator of Notch-driven glioblastoma growth and invasiveness. Cancer Res 2010; 70: 418–427.
Ding X, He Z, Zhou K, Cheng J, Yao H, Lu D et al. Essential role of TRPC6 channels in G2/M phase transition and development of human glioma. J Natl Cancer Inst 2010; 102: 1052–1068.
Yang SL, Cao Q, Zhou KC, Feng YJ, Wang YZ . Transient receptor potential channel C3 contributes to the progression of human ovarian cancer. Oncogene 2009; 28: 1320–1328.
El Boustany C, Bidaux G, Enfissi A, Delcourt P, Prevarskaya N, Capiod T . Capacitative calcium entry and transient receptor potential canonical 6 expression control human hepatoma cell proliferation. Hepatology 2008; 47: 2068–2077.
Kapoor A, Goldberg MS, Cumberland LK, Ratnakumar K, Segura MF, Emanuel PO et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature 2010; 468: 1105–1109.
Novikov L, Park JW, Chen H, Klerman H, Jalloh AS, Gamble MJ . QKI-mediated alternative splicing of the histone variant MacroH2A1 regulates cancer cell proliferation. Mol Cell Biol 2011; 31: 4244–4255.
Sporn JC, Kustatscher G, Hothorn T, Collado M, Serrano M, Muley T et al. Histone macroH2A isoforms predict the risk of lung cancer recurrence. Oncogene 2009; 28: 3423–3428.
Smedlund K, Vazquez G . Involvement of native TRPC3 proteins in ATP-dependent expression of VCAM-1 and monocyte adherence in coronary artery endothelial cells. Arterioscler Thromb Vasc Biol 2008; 28: 2049–2055.
Gifford SM, Yi FX, Bird IM . Pregnancy-enhanced store-operated Ca2+ channel function in uterine artery endothelial cells is associated with enhanced agonist-specific transient receptor potential channel 3-inositol 1,4,5-trisphosphate receptor 2 interaction. J Endocrinol 2006; 190: 385–395.
Gamble MJ, Frizzell KM, Yang C, Krishnakumar R, Kraus WL . The histone variant macroH2A1 marks repressed autosomal chromatin, but protects a subset of its target genes from silencing. Genes Dev 2010; 24: 21–32.
Buschbeck M, Di Croce L . Approaching the molecular and physiological function of macroH2A variants. Epigenetics 2010; 5: 118–123.
Aydar E, Yeo S, Djamgoz M, Palmer C . Abnormal expression, localization and interaction of canonical transient receptor potential ion channels in human breast cancer cell lines and tissues: a potential target for breast cancer diagnosis and therapy. Cancer Cell Int 2009; 9: 23.
Thebault S, Flourakis M, Vanoverberghe K, Vandermoere F, Roudbaraki M, Lehen'kyi V et al. Differential role of transient receptor potential channels in Ca2+ entry and proliferation of prostate cancer epithelial cells. Cancer Res 2006; 66: 2038–2047.
Mietton F, Sengupta AK, Molla A, Picchi G, Barral S, Heliot L et al. Weak but uniform enrichment of the histone variant macroH2A1 along the inactive X chromosome. Mol Cell Biol 2009; 29: 150–156.
Choi J, Kim H, Kim K, Lee B, Lu W, An W . Selective requirement of H2B N-Terminal tail for p14ARF-induced chromatin silencing. Nucleic Acids Res 2011; 39: 9167–9180.
Hokamp K, Roche FM, Acab M, Rousseau ME, Kuo B, Goode D et al. ArrayPipe: a flexible processing pipeline for microarray data. Nucleic Acids Res 2004; 32: W457–W459.
An W, Kim J, Roeder RG . Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 2004; 117: 735–748.
Kim H, Heo K, Choi J, Kim K, An W . Histone variant H3.3 stimulates HSP70 transcription through cooperation with HP1gamma. Nucleic Acids Res 2011; 39: 8329–8341.
Acknowledgements
We thank Dr Edward Seto and Dr Ali Hamiche for the gift of HDAC1/2 and macroH2A1.1 plasmids, respectively. We thank Dr Stefan Dimitrov for anti-macroH2A1 antibody. This work was supported by NIH Grant R01GM84209 and ACS Research Scholar Grant DMC-1005001 awarded to WA.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogenesis website .
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Kim, JM., Heo, K., Choi, J. et al. The histone variant MacroH2A regulates Ca2+ influx through TRPC3 and TRPC6 channels. Oncogenesis 2, e77 (2013). https://doi.org/10.1038/oncsis.2013.40
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/oncsis.2013.40
Keywords
This article is cited by
-
PLAA suppresses ovarian cancer metastasis via METTL3-mediated m6A modification of TRPC3 mRNA
Oncogene (2022)
-
Zebrafish macroH2A variants have distinct embryo localization and function
Scientific Reports (2019)
-
MacroH2A1 chromatin specification requires its docking domain and acetylation of H2B lysine 20
Nature Communications (2018)
-
MacroH2A1.2 inhibits prostate cancer-induced osteoclastogenesis through cooperation with HP1α and H1.2
Oncogene (2018)
-
Variants of core histones and their roles in cell fate decisions, development and cancer
Nature Reviews Molecular Cell Biology (2017)