Abstract
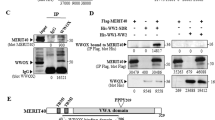
GATA3, a critical transcription factor involved in the development of the mammary gland, also plays important roles in mammary tumorigenesis by regulating transcription in coordination with two essential DNA repair factors, PARP1 and BRCA1. However, whether and how GATA3 participates in the process of DNA repair, which is often associated with tumorigenesis, has not been investigated. Here we demonstrate that GATA3 is required for the repair of DNA double-strand breaks (DSBs) by homologous recominbation (HR). Mechanistic studies indicate that at both the protein and the mRNA level, depleting GATA3 leads to reduced expression of CtIP, an essential HR factor involved in end resection, thereby suppressing the repair of DSBs by HR and sensitizing cells to etoposide induced DNA DSBs. Further studies indicate that upon the occurrence of DNA DSBs GATA3 directly binds to the CtIP promoter at the region of −2119 to −2130 and −2274 to −2285, and promotes the transcription of CtIP. Overexpression of CtIP in GATA3 depleted cells rescues the decline of HR, and cell survival in the presence of etoposide. In addition, through data mining analysis, we observed an extremely strong correlation between the expression levels of GATA3 and CtIP in paratumors, but the correlation turned insignificant in mammary tumors. Using vectors encoding GATA3 with mutations frequently occurring in mammary tumors, we found that several mutations on GATA3 led to a dysregulation of CtIP, and therefore HR repair. In summary, our data delineates the regulatory mechanisms of GATA3 in DNA DSB repair and strongly suggests that it might act as a tumor suppressor by promoting CtIP expression and HR to stabilize genomes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ciocca V, Daskalakis C, Ciocca RM, Ruiz-Orrico A, Palazzo JP . The significance of GATA3 expression in breast cancer: a 10-year follow-up study. Hum Pathol 2009; 40: 489–495.
Pikkarainen S, Tokola H, Kerkela R, Ruskoaho H . GATA transcription factors in the developing and adult heart. Cardiovasc Res 2004; 63: 196–207.
Patient RK, McGhee JD . The GATA family (vertebrates and invertebrates). Curr Opin Genet Dev 2002; 12: 416–422.
Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol 2007; 9: 201–209.
Takaku M, Grimm SA, Wade PA . GATA3 in breast cancer: tumor suppressor or oncogene? Gene Expr 2015; 16: 163–168.
Haveri H, Westerholm-Ormio M, Lindfors K, Maki M, Savilahti E, Andersson LC et al. Transcription factors GATA-4 and GATA-6 in normal and neoplastic human gastrointestinal mucosa. BMC Gastroenterol 2008; 8: 9.
Guo M, Akiyama Y, House MG, Hooker CM, Heath E, Gabrielson E et al. Hypermethylation of the GATA genes in lung cancer. Clin Cancer Res 2004; 10: 7917–7924.
Chou J, Provot S, Werb Z . GATA3 in development and cancer differentiation: cells GATA have it!. J Cell Physiol 2010; 222: 42–49.
Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490: 61–70.
Tkocz D, Crawford NT, Buckley NE, Berry FB, Kennedy RD, Gorski JJ et al. BRCA1 and GATA3 corepress FOXC1 to inhibit the pathogenesis of basal-like breast cancers. Oncogene 2012; 31: 3667–3678.
Shan L, Li X, Liu L, Ding X, Wang Q, Zheng Y et al. GATA3 cooperates with PARP1 to regulate CCND1 transcription through modulating histone H1 incorporation. Oncogene 2014; 33: 3205–3216.
Chen HY, White E . Role of autophagy in cancer prevention. Cancer Prev Res 2011; 4: 973–983.
Aqeilan RI, Croce CM . WWOX in biological control and tumorigenesis. J Cell Physiol 2007; 212: 307–310.
Negrini S, Gorgoulis VG, Halazonetis TD . Genomic instability—an evolving hallmark of cancer. Nat Rev Mol Cell Biol 2010; 11: 220–228.
Davis AJ, Chen BP, Chen DJ . DNA-PK: a dynamic enzyme in a versatile DSB repair pathway. DNA Repair 2014; 17: 21–29.
Zeng-Rong N, Paterson J, Alpert L, Tsao MS, Viallet J, Alaoui-Jamali MA . Elevated DNA repair capacity is associated with intrinsic resistance of lung cancer to chemotherapy. Cancer Res 1995; 55: 4760–4764.
Hsu HM, Wang HC, Chen ST, Hsu GC, Shen CY, Yu JC . Breast cancer risk is associated with the genes encoding the DNA double-strand break repair Mre11/Rad50/Nbs1 complex. Cancer Epidemiol Biomarkers Prev 2007; 16: 2024–2032.
Khanna KK, Jackson SP . DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 2001; 27: 247–254.
Zhang L, Zhang F, Zhang W, Chen L, Gao N, Men Y et al. Harmine suppresses homologous recombination repair and inhibits proliferation of hepatoma cells. Cancer Biol Ther 2015; 16: 1585–1592.
Mao Z, Jiang Y, Liu X, Seluanov A, Gorbunova V . DNA repair by homologous recombination, but not by nonhomologous end joining, is elevated in breast cancer cells. Neoplasia 2009; 11: 683–691.
Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkac J et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell 2013; 49: 872–883.
Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J et al. Human CtIP promotes DNA end resection. Nature 2007; 450: 509–514.
Chen PL, Liu F, Cai S, Lin X, Li A, Chen Y et al. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol Cell Biol 2005; 25: 3535–3542.
Soria-Bretones I, Saez C, Ruiz-Borrego M, Japon MA, Huertas P . Prognostic value of CtIP/RBBP8 expression in breast cancer. Cancer Med 2013; 2: 774–783.
Liu F, Lee WH . CtIP activates its own and cyclin D1 promoters via the E2F/RB pathway during G1/S progression. Mol Cell Biol 2006; 26: 3124–3134.
Huhn D, Kousholt AN, Sorensen CS, Sartori AA . miR-19, a component of the oncogenic miR-17 approximately 92 cluster, targets the DNA-end resection factor CtIP. Oncogene 2015; 34: 3977–3984.
Minchenko DO, Karbovskyi LL, Danilovskyi SV, Moenner M, Minchenko OH . Effect of hypoxia and glutamine or glucose deprivation on the expression of retinoblastoma and retinoblastoma-related genes in ERN1 knockdown glioma U87 cell line. Am J Mol Biol 2012; 2: 21–31.
Mao Z, Seluanov A, Jiang Y, Gorbunova V . TRF2 is required for repair of nontelomeric DNA double-strand breaks by homologous recombination. Proc Natl Acad Sci USA 2007; 104: 13068–13073.
Caron de Fromentel C, Soussi T . TP53 tumor suppressor gene: a model for investigating human mutagenesis. Genes Chromosomes Cancer 1992; 4: 1–15.
Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004; 304: 554.
Eeckhoute J, Keeton EK, Lupien M, Krum SA, Carroll JS, Brown M . Positive cross-regulatory loop ties GATA-3 to estrogen receptor alpha expression in breast cancer. Cancer Res 2007; 67: 6477–6483.
Wu G, Lee WH . CtIP, a multivalent adaptor connecting transcriptional regulation, checkpoint control and tumor suppression. Cell Cycle 2006; 5: 1592–1596.
Kaidi A, Weinert BT, Choudhary C, Jackson SP . Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science 2010; 329: 1348–1353.
Yu X, Chen J . DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol 2004; 24: 9478–9486.
Yu X, Fu S, Lai M, Baer R, Chen J . BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev 2006; 20: 1721–1726.
Jimeno S, Fernandez-Avila MJ, Cruz-Garcia A, Cepeda-Garcia C, Gomez-Cabello D, Huertas P . Neddylation inhibits CtIP-mediated resection and regulates DNA double strand break repair pathway choice. Nucleic Acids Res 2015; 43: 987–999.
Li Z, Zhang W, Chen Y, Guo W, Zhang J, Tang H et al. Impaired DNA double-strand break repair contributes to the age-associated rise of genomic instability in humans. Cell Death Differ 2016; 23: 1765–1777.
Acknowledgements
We thank Dr Michael Van Meter for critically reading the manuscript. We thank Yang Yu for helping with the clonogenic assay. We thank Lu Chen for helping create the vector encoding GATA3. We thank TCGA for providing high quality high-throughput data for public analysis. The work was supported by grants from Chinese National Program on Key Basic Research Project (Grant No. 2013CB967600, 2015CB964800), the National Science Foundation of China (Grant No. 81502385, 81622019, 31371396, 31570813, 81402543, 81601212), the 1000 Youth Talents Program, Shanghai Municipal Natural Science Foundation (Grant No. 15ZR1442600), and the Fundamental Research Funds for the Central Universities.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Zhang, F., Tang, H., Jiang, Y. et al. The transcription factor GATA3 is required for homologous recombination repair by regulating CtIP expression. Oncogene 36, 5168–5176 (2017). https://doi.org/10.1038/onc.2017.127
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2017.127
This article is cited by
-
GATA3 functions downstream of BRCA1 to promote DNA damage repair and suppress dedifferentiation in breast cancer
BMC Biology (2024)
-
Integrated single-cell and bulk RNA sequencing analysis identifies a neoadjuvant chemotherapy-related gene signature for predicting survival and therapy in breast cancer
BMC Medical Genomics (2023)
-
Evaluation of Circ_0000977-Mediated Regulatory Network in Breast Cancer: A Potential Discriminative Biomarker for Triple-Negative Tumors
Biochemical Genetics (2023)
-
Circular RNA hsa_circ_0044234 as distinct molecular signature of triple negative breast cancer: a potential regulator of GATA3
Cancer Cell International (2021)