Abstract
Novel drugs like Abiraterone or Enzalutamide, which target androgen receptor (AR) signaling to improve androgen deprivation therapy (ADT), have been developed during the past years. However, the application of these drugs is limited because of occurrence of inherent or acquired therapy resistances during the treatment. Thus, identification of new molecular targets is urgently required to improve current therapeutic prostate cancer (PCa) treatment strategies. PIAS1 (protein inhibitor of activated STAT1 (signal transducer and activator of transcription-1)) is known to be an important cell cycle regulator and PIAS1-mediated SUMOylation is essential for DNA repair. In this context, elevated PIAS1 expression has already been associated with cancer initiation. Thus, in the present study, we addressed the question of whether PIAS1 targeting can be used as a basis for an improved PCa therapy in combination with anti-androgens. We show that PIAS1 significantly correlates with AR expression in PCa tissue and in cell lines and demonstrate that high PIAS1 levels predict shorter relapse-free survival. Our patient data are complemented by mechanistic and functional in vitro experiments that identify PIAS1 as an androgen-responsive gene and a crucial factor for AR signaling via prevention of AR degradation. Furthermore, PIAS1 knockdown is sufficient to decrease cell proliferation as well as cell viability. Strikingly, Abiraterone or Enzalutamide treatment in combination with PIAS1 depletion is even more effective than single-drug treatment in multiple PCa cell models, rendering PIAS1 as a promising target protein for a combined treatment approach to improve future PCa therapies.
Similar content being viewed by others
Introduction
In recent years, much effort has been made to improve treatment of locally advanced, metastatic and castration-resistant prostate cancer (PCa). Novel drugs like Enzalutamide (formerly MDV3100; Xtandi)1 or Abiraterone (Zytiga),2, 3 which target androgen receptor (AR) signaling to improve first-line androgen deprivation therapy (ADT), or the chemotherapeutic drug Docetaxel (Taxotere), which is given after ADT failure,4, 5 are currently the gold standard treatment options. Although their application resulted in prolonged survival, a single therapy using these drugs is rather limited because of occurrence of an inherent or acquired resistance.6, 7, 8 Recently, it was demonstrated that PCa cells that are resistant to Enzalutamide or Abiraterone not only display cross-resistances to each other, but also to taxanes like Docetaxel.9 This fact further constricts treatment success in the management of PCa and points out the importance for identification of new molecular targets in order to improve future therapeutic strategies.
PIAS (protein inhibitors of activated STAT (signal transducer and activator of transcription), which comprises a family of four multifunctional proteins called PIAS1 to 4, are known to play a significant role in the modulation of various signaling pathways via different molecular mechanisms.10, 11 Besides the DNA and protein binding ability, which is mediated by the conserved SAP domain, PIAS proteins also contain a RING finger-like zinc-binding domain as well as a SUMO interaction motif, thus functioning as SUMO-E3 ligases. Recently, it was demonstrated that PIAS1-mediated SUMOylation is essential for DNA repair.12, 13 Furthermore, PIAS1 is an important cell cycle regulator that promotes cell proliferation by SUMOylation-triggered inhibition of p73 and p53.14, 15, 16 Thus, an increased PIAS1 expression might influence tumor initiation and progression. In this context, a role for PIAS1 in carcinogenesis has already been suggested.17, 18, 19 In line with these findings, we previously reported on significantly elevated PIAS1 levels in primary and metastatic PCa tumors as well as in parental- and docetaxel-resistant tumor cells and suggested an oncogenic role for PIAS1 through regulation of tumor suppressor p21 and the anti-apoptotic protein Mcl-1.20 In addition, it has been demonstrated that PIAS1 can also act as a coregulator of the AR.21, 22 However, the complex relationship between elevated PIAS1 expression and AR signaling in PCa with consequences for PCa progression and patient prognosis has not been investigated so far.
Thus, in the present study, we assess the association of PIAS1 and AR in patient tissue samples and address the question of whether PIAS1 targeting can be used as a basis for an improved PCa therapy in combination with antiandrogens. Patient data are complemented by mechanistic and functional in vitro experiments that identify PIAS1 as an important factor for AR signaling and a promising new target for improved future PCa therapies.
Results
PIAS1 correlates with AR expression in the tissue of PCa patients and cell lines
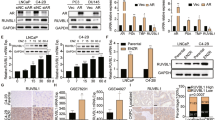
In previous publications20, 23 we have already reported significantly elevated PIAS1 levels in primary tumors and metastatic lesions of treatment-naive and docetaxel-treated PCa patients who had undergone radical prostatectomy, as well as in docetaxel-resistant cell lines. However, the previously used cell lines are AR negative. As the AR signaling cascade is one of the major pathways in PCa that drives tumor progression, the present study aims to elucidate the relationship between PIAS1 and AR signaling. Immunohistochemical analysis of 99 patient samples (Innsbruck cohort) revealed a significant positive correlation between PIAS1 and AR expression (Pearson’s factor 0.674) (Figures 1a and b). Our findings were strengthened by two validation cohorts. The Bonn validation cohort (211 patients; Pearson’s factor 0.591) as well as the Rotterdam validation cohort (443 patients; Pearson’s factor 0.773) confirmed a significant correlation between PIAS1 and AR expression in PCa tissue (Figure 1a). Finally, combining all three cohorts resulted in a total number of 753 PCa patients with Pearson’s correlation coefficient of 0.707 and R2 of 0.50 (Figure 1a; Supplementary Figure S1A). Furthermore, we were able to confirm this correlation in vitro. Screening of AR-positive PCa cells by western blot and immunohistochemistry revealed high PIAS1 as well as AR protein levels in DUCaP and VCaP cells and low PIAS1 and AR expression in LNCaP and CWR22RV1 cells that resulted in Pearson’s correlation factor of 0.843 (Figures 1c and d) and R2 of 0.71 (Supplementary Figure S1B), proving a similar situation in vitro and in PCa tissue.
PIAS1 correlates with AR expression in PCa patients and cell lines and is a marker for reduced relapse-free survival. (a) Pearson’s correlation analysis of PIAS1 and AR immunoreactivity scores (IRS) after immunohistochemistry (IHC) staining of tumor tissue samples from three independent patient cohorts (Innsbruck, Bonn and Rotterdam cohorts). Correlation analysis after pooling all three cohorts (n=753) resulted in R=0.773 (P-value 3.5E−115). (b) Representative benign and malignant tissue cores of two patients with low and high PIAS1/AR expression (scale bar=100 μm). (c) Pearson’s correlation analysis of PIAS1 and AR expression in DUCaP, VCaP and CWR22RV1 cells (R=0.843; P-value 0.001). Western blot analysis for PIAS1 and AR expression was performed in three independent experiments and was used for the correlation analysis. (d) IHC staining of PIAS1 and AR in embedded LNCaP, DUCaP, VCaP and CWR22RV1 cells (scale bar=100 μm). (e) Survival analysis was performed using 10-year follow-up data from 735 patients from all three cohorts. Kaplan–Maier statistics (log rank (Mantel–Cox); P-value: 0.011) of relapse-free survival (defined as time to prostate serum antigen (PSA) progression) of patients with low PIAS1 expression (IRS ≤4) versus patients with intermediate or high PIAS1 expression (IRS >4).
High PIAS1 expression is a marker for decreased relapse-free survival
Strikingly, by combining PIAS1 immunoreactivity score with the pathological background data of patients (n=735) from all three patient cohorts, we were able to identify PIAS1 as a marker for biochemical relapse (defined as rising prostate serum antigen levels) (Figure 1e). Kaplan–Maier statistics revealed that patients with a low PIAS1 immune reactivity score (immunoreactivity score ≤4) display a prolonged biochemical relapse-free survival when compared with patients with intermediate or high PIAS1 expression (immunoreactivity score >4; log rank (Mantel–Cox); P-value: 0.011). However, no significant difference in relapse-free survival could be observed in patients with low AR expression compared with those who display intermediate or high AR expression (log rank (Mantel–Cox); P-value: 0.101; Supplementary Figure S1C). Taken together, we conclude from these findings that (1) PIAS1 expression significantly correlates with AR expression in primary tumors and (2) high PIAS1 expression is an indicator for shorter progression-free survival, thereby suggesting an essential role for PIAS1 during PCa progression.
PIAS1 expression is upregulated by androgens at mRNA and protein levels
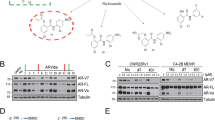
Given that our data both from patient material and cell lines revealed a significant correlation between PIAS1 and AR expression, we next wanted to evaluate the influence of androgens on PIAS1. Thus, we treated LNCaP cells with increasing concentrations of the synthetic androgen R1881 in the absence or presence of the antiandrogen Bicalutamide for 24 h. Androgen treatment induced a dose-dependent significant increase in PIAS1 mRNA and protein expression that could be reversed by Bicalutamide (Figure 2a). Next, we performed a time-course experiment and treated LNCaP, DUCaP and VCaP cells with 1 nM R1881 for 8, 24, 48 and 72 h and evaluated PIAS1 mRNA and protein expression, respectively. In all three cell lines, PIAS1 mRNA peaked after 8–24 h, whereas PIAS1 protein expression was maximally induced after 24–48 h (Figures 2b and c). Immunofluorescence staining confirmed an increased AR expression and nuclear translocation as well as elevated PIAS1 levels upon androgen treatment (Figure 2d). To exclude any unspecific effects of the treatment, the AR-negative cell lines PC3 and DU145 were treated with R1881 for 24 h but no change in PIAS1 mRNA or protein expression was detectable (Supplementary Figure S2A). In addition, to prove androgenic regulation of PIAS1, we performed short-term AR knockdown using two specific AR short interfering RNAs (siRNAs; siAR-1 and siAR-2), resulting in a significant decline of PIAS1 protein (~50% downregulation) in DUCaP as well as in LNCaP cells (Figure 2e).
PIAS1 expression is regulated by androgens at mRNA and protein levels. (a) Cells were treated with increasing concentrations of R1881 (0.1, 0.5, 1 and 10 nM) in the absence or presence of 5 μM Bicalutamide and subjected to real-time quantitative reverse transcription PCR (qRT-PCR) or western blot. Data represent mean+s.e.m. from at least three independent experiments (***P<0.001). (b, c) PIAS1 mRNA and protein expression in LNCaP, DUCaP and VCaP cells after stimulation with 1 nM R1881 for 8, 24, 48 or 72 h. Data represent mean+s.e.m. from three independent experiments (*P<0.05; **P<0.01; ***P<0.001). (d) Immunofluorescence staining for PIAS1 (red) and AR (green) after treatment of LNCaP, DUCaP and VCaP cells with 1 nM R1881 for 24 h. Scale bar=50 μm. (e) Western blot analysis of PIAS1 and AR in DUCaP and LNCaP cells following transfection with two specific AR siRNAs (siAR-1 and siAR-2; 25 nM). Quantifications of western blots represent mean+s.e.m. from at least three independent experiments (*P<0.05; ***P<0.001).
PIAS1 regulation by androgens is a direct transcriptional effect
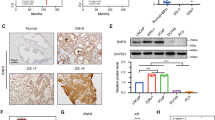
We next aimed to assess whether PIAS1 regulation by androgens is a direct, transcriptional effect or an indirect mechanism. We therefore compared the velocity of PIAS1 mRNA induction upon R1881 treatment with that of FKBP5, which is known to be a direct target gene of AR. A time-course experiment demonstrated that FKBP5 as well as PIAS1 are rapidly upregulated upon treatment, showing a significant induction after 2–4 h in DUCaP (Figure 3a) and LNCaP cells (Supplementary Figure S3A), indicating a similar direct mechanism of AR action for PIAS1 and FKBP5. In concordance with this hypothesis, an AR chromatin immunoprecipitation sequencing (ChIP-Seq) analysis previously performed by our group in DUCaP cells24 revealed several AR-binding sites upstream of the PIAS1 gene following androgen stimulation (Figure 3b), with one binding site (binding site 4) located 11 269 bp upstream to the transcription start site in the PIAS1 promoter region. The in silico analysis of the respective genomic sequence using the Encode Integrated Regulation Track (http://nar.oxfordjournals.org/content/41/D1/D56.long) available in UCSC browser (http://genome.cshlp.org/content/12/6/996.abstract) identified a region that is frequently occupied by transcription factors (Supplementary Figure S2B). In addition, JASPAR database (http://nar.oxfordjournals.org/content/early/2013/11/04/nar.gkt997.full) predicted five androgen response elements within this region (Supplementary Figure S2B). Taken together, these data strongly indicate that PIAS1 upregulation by androgens is a direct transcriptional event.
PIAS1 binds and stabilizes AR and protects AR from proteasomal degradation. (a) Real-time quantitative reverse transcription PCR (qRT-PCR) analysis of DUCaP after treatment with R1881 (1 nM) for different durations, showing velocity of PIAS1 and FKBP5 mRNA transcription following AR activation. Data represent mean+s.e.m. from three independent experiments. (b) Analysis of ChIP-Seq data of DUCaP cells that were treated with vehicle or 1 nM of R1881 for 1 h24 shows AR enrichment sites in close proximity of the PIAS1 gene. (c) Western blot for AR and PIAS1 following co-immunoprecipitation of flag-PIAS1 and endogenous AR in DUCaP after transfection of 1 μg pFlag-PIAS1 or empty vector (EV) for 3 days in the absence or presence of R1881. (d) Western blot for AR and PIAS1 expression in DUCaP after transfection with control siRNA (neg.C) or PIAS1 siRNAs. Data represent mean+s.e.m. from three independent experiments (***P<0.001). (e) Velocity of AR degradation was measured in a time-course experiment by western blot after protein synthesis inhibition with cycloheximide (25 μg/ml) in control- or siPIAS1-transfected DUCaP cells. Data represent mean+s.e.m. from three independent experiments (*P<0.05 siPIAS1-1, siPIAS1-3 at 3 and 5 h; *P<0.05 siPIAS1-3 at 8 h). (f) Western blot for AR and PIAS1 in DUCaP cells that were transfected with control siRNA or PIAS1 siRNA for 72 h and subsequently treated with MG132 (25 μg/ml) for 8 h.
PIAS1 binds and stabilizes AR and protects AR from proteasomal degradation
As PIAS1 expression was found to be androgen regulated and PIAS1 is an identified AR coactivator,21 we next wanted to prove a direct interaction of PIAS1 and the AR by co-immunoprecipitation. Using a flag-tagged PIAS1 expression vector we confirmed specific binding of PIAS1 to endogenous AR in DUCaP and LAPC4 cells (Supplementary Figure S3B). Furthermore, treatment with R1881 resulted in an increased amount of PIAS1-AR complexes (Figure 3c). Next, we wanted to evaluate whether PIAS1 influences AR expression. Short-term PIAS1 knockdown using two specific PIAS1 siRNAs (siPIAS1-1 and siPIAS1-3) resulted in a significant decline of AR protein (~50% downregulation) in DUCaP cells (Figure 3d). It is known that PIAS1 binding to target proteins may influence protein stability.15 Thus, we hypothesized that reduced AR levels in the absence of PIAS1 are a result of accelerated AR degradation. In order to verify this hypothesis, we assessed AR decay in control- or siPIAS1-treated DUCaP cells along a time course after adding the protein synthesis inhibitor cycloheximide. We observed an increased velocity of AR degradation in cells where PIAS1 was depleted (Figure 3e). As a control, treatment with the proteasome inhibitor MG132 caused an increase in AR levels even in the absence of PIAS1 (Figure 3f), proving that the proteasome accounts for the observed effect. These results clearly demonstrate that PIAS1 delays the proteasomal degradation of AR and is therefore a critical factor for AR protein stability.
PIAS1 boosts AR transcriptional activity and expression of AR targets
Having shown that PIAS1 is important for AR protein stability, we next aimed to verify a possible influence of PIAS1 on AR transcriptional activity. Luciferase reporter assays (normalized to cell number/protein content) after PIAS1 knockdown and R1881 treatment revealed significantly decreased AR activity in DUCaP and LNCaP cells, in which PIAS1 was downregulated (Figure 4a). Functionally, the observed decline in AR activity had also consequences on AR target gene expression. PIAS1 knockdown resulted in reduced KLK3 mRNA expression in both investigated cell lines (Figure 4b). However, overexpression of wild-type PIAS1 neither in the absence nor in the presence of R1881 caused significant changes in KLK3 mRNA levels. Summarizing these results, we propose that PIAS1 is important for AR protein stability and activity with consequences for the expression of at least a subgroup of AR downstream targets.
PIAS1 is important for AR activation, influences AR downstream targets and prolonged PIAS1 knockdown results in reduced cell proliferation, viability and increased apoptosis. (a) AR activity was measured using luciferase reporter assays performed after transfection with control siRNA (neg.C) or PIAS1 siRNAs (72 h; 25 nM) and subsequent R1881 treatment (1 nM; last 24 h). Data represent mean+s.e.m. from at least three independent experiments (*P<0.05; ***P<0.001). (b) Real-time quantitative reverse transcription PCR (qRT-PCR) for PIAS1 and KLK3 after transfection with PIAS1 siRNAs, PIAS1 expression vector (PIAS1 WT) or the respective controls. Data represent mean+s.e.m. from three independent experiments (*P<0.05; **P<0.01). (c–e) Proliferation ([3H]thymidine), viability (WST) and apoptosis (flow cytometry) measurements of LNCaP and DUCaP after prolonged (6 days) PIAS1 knockdown (*P<0.05; **P<0.01; ***P<0.001).
PIAS1 knockdown in combination with Enzalutamide and Abiraterone is more effective than single drug treatment
As PIAS1 seems to be a critical factor for AR stability and activity, we next aimed to evaluate a possible effect on PCa cell proliferation and survival after prolonged (6 days) PIAS1 knockdown. [3H]thymidine incorporation assays revealed a significantly decreased proliferation (Figure 4c). WST and sub-G1 measurements uncovered a significantly reduced cell viability and increased apoptosis in DUCaP and LNCaP cells where PIAS1 levels have been depleted (Figures 4d and e). These results clearly indicate that PIAS1 is a critical factor for proliferation and survival of AR-positive PCa cells and might therefore be a promising candidate for a combination treatment with antiandrogens. To test this hypothesis we next performed single and combined treatments with the antiandrogens Abiraterone and Enzalutamide as well as PIAS1 knockdown. As expected, all single treatments (PIAS1 knockdown, Abiraterone and Enzalutamide) resulted in significantly reduced proliferation and cell viability in LNCaP cells and to a lesser extent in DUCaP cells. A possible explanation for the lower sensitivity of DUCaP cells to antiandrogenic drugs might be the increased AR expression in this cell line compared with LNCaP cells (see Figure 1c). Strikingly, combined treatment with either Abiraterone and PIAS1 knockdown or Enzalutamide and PIAS1 knockdown was superior compared with all single treatment approaches as revealed by [3H]thymidine incorporation and WST measurements (Figures 5a and b). Representative pictures of all treatment groups (Figure 5c) showed, in general, less cells and a more apoptotic phenotype of the remaining cells in the combination treatment groups, thus confirming the results obtained in proliferation assays. In addition, we observed elevated p21 mRNA expression within all single treatments, and this was further increased in the combined treatment groups (Supplementary Figure S3C).
PIAS1 knockdown in combination with the antiandrogens Enzalutamide or Abiraterone is superior compared with single drug treatment. (a) [3H]thymidine and (b) WST assay of DUCaP and LNCaP cells that were transfected with ctrl siRNA (neg.C) or PIAS1 siRNA (siPIAS1-1 and siPIAS1-3) twice for 6 days. At 4 h after the transfections, cells were treated with 2.5 μM Enzalutamide (Enza) or Abiraterone (Abi), respectively. Ethanol was used as vehicle for both drugs. Data represent mean+s.e.m. from at least three independent experiments (*P<0.05; **P<0.01; ***P<0.001). (c) Representative light microscopy images for all treatments in DUCaP and LNCaP cells (magnification × 40).
PIAS1 knockdown results in reduced cell viability in Bicalutamide-resistant and AR-negative LNCaP sublines
To further evaluate the potential use of PIAS1 knockdown for treatment of cells that are resistant to antiandrogens or lack AR, we performed single and combination treatments as described above with LNCaP-Bic and LNCaP-IL6+ cells that were established to mimic a castration-resistant cancer cell phenotype. Morphology of all cell lines as well as AR and PIAS1 expression were examined (Supplementary Figures S4A and B). Treatment with 2.5 μM Bicalutamide had no effect on proliferation and cell viability in LNCaP-Bic cells. In contrast, PIAS1 knockdown with both siRNAs resulted in reduced proliferation and cell viability. Treatment with the novel antiandrogens Abiraterone and Enzalutamide reversed Bicalutamide resistance and resulted in reduced proliferation and cell viability as single treatment. Strikingly, PIAS1 knockdown in combination with Abiraterone or Enzalutamide caused an additive inhibitory effect on proliferation and viability (Figures 6a and b, Supplementary Figure S4C). Similar results could be observed when LNCaP-IL6+ cells were used. As these cells lack AR, neither Abiraterone nor Enzalutamide treatment was effective. However, PIAS1 knockdown was sufficient to significantly reduce cell proliferation and viability (Supplementary Figure S5), demonstrating additional mechanisms for PIAS1 action beyond AR.
PIAS1 knockdown sensitizes Bicalutamide-resistant PCa cells to apoptosis. (a) [3H]thymidine and (b) WST assay of LNCaP-Bic cells that were transfected with ctrl siRNA (neg.C) or PIAS1 siRNA (siPIAS1-1 and siPIAS1-3) twice for 6 days. At 4 h after the transfections, cells were treated with 2.5 μM Bicalutamide (Bic), Enzalutamide (Enza) or Abiraterone (Abi), respectively. Ethanol was used as vehicle for both drugs. Data represent mean+s.e.m. from at least three independent experiments (*P<0.05; **P<0.01; ***P<0.001). (c) Proposed mechanism of PIAS1 and AR interaction: stimulation with dihydrotestosterone (DHT) leads to AR activation, translocation and binding to androgen response elements on DNA, resulting in transcription of androgen-responsive genes and PIAS1. PIAS1 in turn binds AR that keeps the receptor in an active state and prevents its proteasomal degradation. As shown in a previous study, PIAS1 acts in addition as a suppressor of p21 in PCa cells, thereby further increasing cellular proliferation.
Discussion
We previously demonstrated that PIAS1 expression increases with PCa malignancy and influences cell proliferation and apoptosis through p21 and Mcl-1 regulation.20, 23 Interestingly, it has also been demonstrated that PIAS1 can act as a coregulator of AR.21, 22 However, the mechanistic background of elevated PIAS1 expression in PCa and the consequences for PCa tumor progression and patient prognosis have not been investigated so far.
Herein, we prove that PIAS1 itself is significantly upregulated upon androgenic stimulation at both mRNA and protein levels in a dose-dependent manner. This result is in line with findings by Heemers et al.25 who showed increased PIAS1 mRNA in LNCaP cells upon androgen treatment. Considering an elevated AR signaling in PCa, this finding might provide one mechanistic explanation for increased PIAS1 levels in PCa patients and the observed association of PIAS1 expression with tumor malignancy described recently by our group.20, 23 In concordance with this hypothesis, in the present study we demonstrate a highly significant, positive correlation of PIAS1 and AR expression in tissue samples from three independent patient cohorts with a total of 753 patient samples, as well as in AR-positive PCa cell lines. Strikingly, we also prove that patients with high PIAS1 levels are significantly more susceptible to biochemical recurrence within 10 years after radical prostatectomy. Interestingly, despite strong correlation of PIAS1 and AR in tumor tissue samples, high AR expression was not found to be predictive for decreased progression-free survival, indicating that PIAS1 is the determining factor for the observed effect on patient survival.
In cell culture experiments we prove that PIAS1 protein directly binds to endogenous AR and that PIAS1-bound AR is slightly more abundant after androgen treatment. However, even under steroid-depleted conditions we were able to detect AR binding to the nuclear protein PIAS1. This result is in concordance with a recent publication demonstrating PIAS1 binding to AR in VCaP cells in the absence or presence of R1881 and might be explained by the fact that AR is partly also present in the nuclei in steroid-depleted cells (Figure 2d.26 Functional studies show that PIAS1 binding to AR causes stabilization of the receptor, thus supporting AR activity. As a consequence, downregulation of PIAS1 in the presence of androgens decreased AR transcriptional activity.
On the basis of these facts, we propose the following mechanism (Figure 6c). The presence of dihydrotestosterone leads to AR activation, nuclear translocation and binding to androgen response elements. Subsequently, transcription of androgen-responsive genes such as prostate serum antigen and PIAS1 is enhanced. PIAS1 in turn binds AR protein and impairs its proteasomal degradation, which keeps the receptor in an active state, thereby supporting AR signaling. However, two open questions remain. First, it is not completely clarified yet which mechanism is responsible for PIAS1 induction upon androgenic stimulation. PIAS1 might be upregulated either directly via AR binding to the PIAS1 promoter or indirectly via other signaling molecules that are themselves influenced by androgens. Nevertheless, the fact that PIAS1 is induced as rapid (2–4 h after R1881 treatment) as the known direct AR downstream target FKBP5 indicates a direct, transcriptional effect of AR on PIAS1. In concordance with this hypothesis, AR ChIP-Seq analysis revealed several AR-binding sites upstream of the PIAS1 gene following androgen stimulation. The in silico analysis of the respective genomic sequence identified a region that is frequently occupied by transcription factors and harbors five predicted androgen response elements. These facts strongly indicate that PIAS1 is a direct AR downstream target that is upregulated by androgens.
Second, the mechanism by which PIAS1 stabilizes AR needs to be further clarified. In this context, it has already been shown that AR is target of SUMOylation.27 The consequences of AR SUMOylation are currently not completely clear. A recent publication reported that SUMO modification might influence AR transcriptional activity positively or negatively in a target gene- and pathway-selective manner.28 In addition, SUMOylation has been shown to define AR half-life in the nucleus, given that a SUMOylation-deficient AR mutant exhibited a decreased lifespan compared with wild-type AR.29, 30 The results obtained in the present study support this view, demonstrating that knockdown of the SUMO E3 ligase PIAS1 increased the proteasomal degradation of AR. Of note, elevated AR expression and/or stability were shown to sensitize AR to low levels of androgens and possibly lead to castration resistance.31 Considering these facts, we hypothesize that high PIAS1 expression might be involved in the development of CRPC via SUMOylation-dependent prevention of AR degradation. However, the exact role of PIAS1 in the development of castration resistance needs to be further investigated.
In addition to supporting AR activity via protein stabilization, PIAS1 has recently been shown to act as a genuine, chromatin-bound AR coregulator that affects AR signaling positively or negatively in a target gene-selective manner.22 Transcriptome profiling analyses performed by Toropainen et al.22 indicate that PIAS1 depletion affects one-tenth of all androgen-regulated genes, among them predominantly genes involved in proliferation and cell cycle regulation. In addition, a subset of genes becomes subjected to androgen regulation whereas other genes completely lose androgen responsiveness in the absence of PIAS1. The mechanism underlying these effects is suggested to be at least in part SUMOylation of FOXA1. Thus, PIAS1 does not simply act as a general AR co-activator but has a more comprehensive role in defining a specific transcriptional program relevant to PCa cell growth upon androgen treatment.
In the past years, much effort has been made in order to improve ADT. Novel drugs like Abiraterone and Enzalutamide are promising, but the therapeutic efficacy varies.32, 33 Herein, we investigated the potential of PIAS1 inhibition to improve current PCa treatment strategies. We found that PIAS1 knockdown increases the efficacy of ADT drugs and, most importantly, induces growth arrest and apoptosis even as a single treatment, irrespective of the cell’s AR status or sensitivity to the antiandrogen Bicalutamide. Interestingly, we found increased p21 levels not only after PIAS1 depletion, but also after single treatment with Abiraterone or Enzalutamide. The use of either drug in combination with PIAS1 knockdown further increased p21 levels. Further studies may dissect the role of p21 in the responsiveness or resistance to Enzalutamide and Abiraterone.
In conclusion, the presented data provide the rationale for clinical PIAS1 targeting either alone or in combination with novel ADT drugs, such as Abiraterone or Enzalutamide.
Materials and methods
Cell culture and chemicals
LNCaP, DUCaP, VCAP, CWR22RV1 and LAPC4 cells were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). LNCaP, DUCaP, CWR22RV1 and LAPC4 cells were cultured in RPMI-1640 supplemented with 10% fetal calf serum and 2 mM glutamax (Fisher Scientific, Vienna, Austria). LAPC4 cells were further supplemented with 100 nM dihydrotestosterone. VCaP cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 4 mM glutamax and 1.75 g D-glucose. LNCaP-IL6+ cells were generated after long-term exposure to IL6 as described elsewhere 34. LNCaP-Bic cells are resistant to Bicalutamide and were obtained after long-term treatment of LNCaP cells with Bicalutamide as reported earlier.35 Identity of the used cell lines was confirmed by short tandem repeat analysis. Experiments using the synthetic androgen R1881 were performed in media containing 10% charcoal-stripped fetal calf serum (GE Healthcare, Vienna, Austria). For protein stability analysis, cells were treated with 25 μg/ml cycloheximide (Sigma, Vienna, Austria) or 25 μg/ml MG132 (Sigma) and harvested after different incubation times. The lysates were then subjected to western blot. For single and combined drug treatments, cells were transfected with 25 nM of control or PIAS1 siRNA and then treated with 2.5 μM of Abiraterone, Enzalutamide or Bicalutamide, respectively.
Tissue microarray (TMA) and immunohistochemistry
In the Innsbruck cohort, TMA processing and immunohistochemistry evaluation of tissue samples obtained from 99 patients and 4 cell lines were performed as described previously.20, 36 The use of archived material was approved by the Ethics Committee of Medical University of Innsbruck (Study no. AM 3174 including amendment 2).
In the Rotterdam cohort, use of samples was approved by the Erasmus Medical Center Medical Ethics Committee according to the Medical Research involving Human Subjects Act (MEC-2011-295 and MEC-2011-296). TMA was constructed from 481 PCa patients as described in detail elsewhere.37
In the Bonn cohort, a TMA was constructed from 237 patients with localized prostate adenocarcinoma who underwent radical prostatectomy at the University Hospital Bonn between 2000 and 2008. The study has been approved by the institutional review board at the University Hospital Bonn (Lfd. Nr. 071/14).
The following antibodies were used: anti-PIAS1 (1:400; ab77321, Abcam, Cambridge, UK) and anti-AR (1:200; sc-816, Santa Cruz Biotechnology, Santa Cruz, CA, USA). TMAs were evaluated using the following modified ‘quick-score’ protocol: staining intensity was scored 0–3 (0=absent, 1=weak, 2=intermediate, 3=strong). The percentage of positively stained cells was scored 0–4 (0=absent, 1=<10%, 2=<50%, 3=<75%, 4=>75%). Both scores were multiplied to obtain an immunoreactivity score, ranging from 0 to 12.
Immunofluorescence
Cells were seeded onto glass coverslips and immunofluorescence was performed as previously described.20 The following antibodies were used: anti-PIAS1 (1:500; 3550S, Cell Signaling, Danvers, MA, USA), anti-AR (1:100; sc-816, Santa Cruz Biotechnology), secondary antibody goat-anti-rabbit 555 (10082602, Life Technologies Carlsbad, CA, USA) and donkey-anti-mouse 488 (A21202, Life Technologies). Experiments were performed in three biological replicates.
siRNA transfection
The following siRNA sequences were used for targeting human PIAS1 and AR: siPIAS1-1: 5′-AAGGUCAUUCUAGAGCUUUAdTdT-3′, siPIAS1-3: 5′-CGAAUGAACUUGGCAGAAAdTdT-3′, siAR-1: 5′-GCACUGCUACUCUUCAGCAdTdT-3′ and siAR-2: 5′-GACCUACCGAGGAGCUUUCdTdT-3′. A nontargeting siRNA pool (Cat. no. D-001810-10-20, Dharmacon, Chicago, IL, USA) was used as a negative control. siRNA transfections were performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) reagent according to the manufacturer’s protocol. All cell lines were transfected with either 25 nM of siRNAs against PIAS1, AR or nontargeting control. To ensure prolonged knockdown of the target proteins for 6 days, all cell lines were re-transfected with the same concentration of the respective siRNA at day 3.
Plasmids
Expression vectors pEGFP-C1-PIAS1 wild-type and empty control vector were generated by Dr Yaron Galanty (Gurdon Institute, Cambridge, UK) as described elsewhere.13 For Flag PIAS1 overexpressing construct, PIAS1 wild type was cloned in a pDest-Flag control vector as used in Mikolcevic et al.38 Cells were transfected with 3 μg per 6-well of DNA using X-tremeGENE HP transfection reagent (Roche, Basel, Switzerland) for 72 h following the manufacturer’s instructions.
Real-time quantitative reverse transcription PCR
RNA isolation, complementary DNA synthesis, and real-time quantitative reverse transcription PCR were performed as described elsewhere.39 Expression was normalized to the endogenous reference TATA-Box-binding protein (forward 5′-CACGAACCACGGCACTGATT-3′; reverse 5′-TTTTCTGCTGCCAGTCTGGAC-3′; probe 5-FAM-TCTTCACTCTTGGCTCCTGTGCACA-TAMRA-3). PIAS1, AR, KLK3 and p21 Taqman gene expression assays, purchased from Applied Biosystems (Foster City, CA, USA), were used according to the manufacturer’s protocol. Real-time quantitative reverse transcription PCR experiments were done in at least three independent biological experiments with three technical replicates.
Western blot
Western blot was performed as previously described.39 The following antibodies were used: anti-GAPDH (1:100 000; MAB374, Millipore, Vienna, Austria), anti-AR (1:500; sc-816, Santa Cruz Biotechnology), anti-PIAS1 (1:500; 3550S, Cell Signaling). All western blots were performed in at least three independent biological experiments.
Proliferation, viability and apoptosis measurement
Proliferation was assessed using [3H]thymidine incorporation as previously described.39 For viability measurement, WST assay (Roche) was performed according to the manufacturer’s protocol. The percentage of apoptotic cells was assessed by flow cytometry as previously described.20 For all assays, cells were transfected twice within a period of 6 days. Measurements were done in at least three independent biological experiments with three technical replicates.
Dual-luciferase reporter assay
LNCaP and DUCaP were transfected with 0.5 μg pGL3-ARE2TATA reporter plasmid (Firefly luciferase), 0.06 μg pGL4.73 control reporter plasmid (Renilla luciferase) and 50 nM of the respective siRNA using Lipofectamine 2000 reagent. At day 4, cells were treated with 1 nM R1881 for 24 h, followed by cell lysis and quantification of luciferase activity using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). Luciferase activity was assessed using the Chameleon 5025 (HVD Life Sciences, Vienna, Austria) and firefly luciferase activity was normalized to Renilla luciferase activity. The values were then normalized to total protein content. Luciferase assays were done in at least three independent biological experiments with three technical replicates.
Co-immunoprecipitation
Cells were transfected with 1 μg of pFlag-PIAS1 or empty vector. Cells were lyzed in buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, protease inhibitor (Thermo Scientific, Waltham, MA, USA) and phosphatase inhibitor (Sigma). PIAS1 conjugates were precipitated by incubating lysates with Anti-FLAG M2 Magnetic Beads (Sigma) for 2 h. Samples were washed 2 times with lysis buffer and 3 times with TBS and subsequently eluted with LDS sample buffer at 70 °C for 10 min. Lysates were subjected to western blot under reducing conditions. The following antibodies were used: anti-AR (1:2000; sc-816, Santa Cruz Biotechnology), anti-PIAS1 (1:2000; 3550S, Cell Signaling), goat anti rabbit HRP (111-035-003, Jackson ImmunoResearch, Westgrove, PA, USA). Co-immunoprecipitation experiments were performed as three independent biological experiments.
ChIP-Seq data
ChIP has been published previously.24 Briefly, DUCaP cells were treated with vehicle or 1 nM R1881 for 1 h and precipitated DNA was analyzed by deep sequencing.
Statistical analysis
SPSS (V15.0, IBM, Armonk, NY, USA) and GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA) were used for statistical analyses. For all experiments, Gaussian distribution was determined using Kolmogorov–Smirnov test. Differences between treatment groups were analyzed using Student’s t-test or Mann–Whitney U-test and were corrected for multiple testing using Bonferroni method. Correlation analysis was performed by the Pearson’s method. Differences in recurrence-free survival were assessed using Kaplan–Meier plots and log-rank test. Two consecutive prostate serum antigen measurements >0.2 ng/ml were considered as biochemical recurrence. P-values of <0.05 were considered significant. All differences highlighted by asterisks were statistically significant as encoded in figure legends (*P<0.05, **P<0.01, ***P<0.001). Data are presented as mean+s.e.m. unless otherwise specified.
References
Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med 2012; 367: 1187–1197.
Ryan CJ, Smith MR, de Bono JS, Molina A, Logothetis CJ, de Souza P et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med 2013; 368: 138–148.
de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med 2011; 364: 1995–2005.
Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med 2004; 351: 1502–1512.
Petrylak DP, Tangen CM, Hussain MH, Lara Jr PN, Jones JA, Taplin ME et al. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N Engl J Med 2004; 351: 1513–1520.
Geney R, Ungureanu LM, Li D, Ojima I . Overcoming multidrug resistance in taxane chemotherapy. Clin Chem Lab Med 2002; 40: 918–925.
Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res 2012; 72: 3457–3462.
Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM . Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res 2013; 73: 483–489.
van Soest RJ, van Royen ME, de Morree ES, Moll JM, Teubel W, Wiemer EA et al. Cross-resistance between taxanes and new hormonal agents abiraterone and enzalutamide may affect drug sequence choices in metastatic castration-resistant prostate cancer. Eur J Cancer 2013; 49: 3821–3830.
Sharrocks AD . PIAS proteins and transcriptional regulation—more than just SUMO E3 ligases? Genes Dev 2006; 20: 754–758.
Rytinki MM, Kaikkonen S, Pehkonen P, Jaaskelainen T, Palvimo JJ . PIAS proteins: pleiotropic interactors associated with SUMO. Cell Mol Life Sci 2009; 66: 3029–3041.
Shima H, Suzuki H, Sun J, Kono K, Shi L, Kinomura A et al. Activation of the SUMO modification system is required for the accumulation of RAD51 at sites of DNA damage. J Cell Sci 2013; 126: 5284–5292.
Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP . Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 2009; 462: 935–939.
Kahyo T, Nishida T, Yasuda H . Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol Cell 2001; 8: 713–718.
Munarriz E, Barcaroli D, Stephanou A, Townsend PA, Maisse C, Terrinoni A et al. PIAS-1 is a checkpoint regulator which affects exit from G1 and G2 by sumoylation of p73. Mol Cell Biol 2004; 24: 10593–10610.
Schmidt D, Muller S . Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc Natl Acad Sci USA 2002; 99: 2872–2877.
Liu B, Tahk S, Yee KM, Yang R, Yang Y, Mackie R et al. PIAS1 regulates breast tumorigenesis through selective epigenetic gene silencing. PLoS One 2014; 9: e89464.
Li R, Wei J, Jiang C, Liu D, Deng L, Zhang K et al. Akt SUMOylation regulates cell proliferation and tumorigenesis. Cancer Res 2013; 73: 5742–5753.
Driscoll JJ, Pelluru D, Lefkimmiatis K, Fulciniti M, Prabhala RH, Greipp PR et al. The sumoylation pathway is dysregulated in multiple myeloma and is associated with adverse patient outcome. Blood 2010; 115: 2827–2834.
Hoefer J, Schafer G, Klocker H, Erb HH, Mills IG, Hengst L et al. PIAS1 is increased in human prostate cancer and enhances proliferation through inhibition of p21. Am J Pathol 2012; 180: 2097–2107.
Gross M, Liu B, Tan J, French FS, Carey M, Shuai K . Distinct effects of PIAS proteins on androgen-mediated gene activation in prostate cancer cells. Oncogene 2001; 20: 3880–3887.
Toropainen S, Malinen M, Kaikkonen S, Rytinki M, Jaaskelainen T, Sahu B et al. SUMO ligase PIAS1 functions as a target gene selective androgen receptor coregulator on prostate cancer cell chromatin. Nucleic Acids Res 2014; 43: 848–861.
Puhr M, Hoefer J, Neuwirt H, Eder IE, Kern J, Schafer G et al. PIAS1 is a crucial factor for prostate cancer cell survival and a valid target in docetaxel resistant cells. Oncotarget 2014; 5: 12043–12056.
Bu H, Schweiger MR, Manke T, Wunderlich A, Timmermann B, Kerick M et al. Anterior gradient 2 and 3—two prototype androgen-responsive genes transcriptionally upregulated by androgens and by oestrogens in prostate cancer cells. FEBS J 2013; 280: 1249–1266.
Heemers HV, Regan KM, Schmidt LJ, Anderson SK, Ballman KV, Tindall DJ . Androgen modulation of coregulator expression in prostate cancer cells. Mol Endocrinol 2009; 23: 572–583.
Guo Z, Dai B, Jiang T, Xu K, Xie Y, Kim O et al. Regulation of androgen receptor activity by tyrosine phosphorylation. Cancer Cell 2006; 10: 309–319.
Poukka H, Karvonen U, Janne OA, Palvimo JJ . Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1). Proc Natl Acad Sci USA 2000; 97: 14145–14150.
Sutinen P, Malinen M, Heikkinen S, Palvimo JJ . SUMOylation modulates the transcriptional activity of androgen receptor in a target gene and pathway selective manner. Nucleic Acids Res 2014; 42: 8310–8319.
Rytinki M, Kaikkonen S, Sutinen P, Paakinaho V, Rahkama V, Palvimo JJ . Dynamic SUMOylation is linked to the activity cycles of androgen receptor in the cell nucleus. Mol Cell Biol 2012; 32: 4195–4205.
van der Steen T, Tindall DJ, Huang H . Posttranslational modification of the androgen receptor in prostate cancer. Int J Mol Sci 2013; 14: 14833–14859.
Feldman BJ, Feldman D . The development of androgen-independent prostate cancer. Nat Rev Cancer 2001; 1: 34–45.
Heidegger I, Nagele U, Pircher A, Pichler R, Horninger W, Bektic J . Latent hypothyreosis as a clinical biomarker for therapy response under abiraterone acetate therapy. Anticancer Res 2014; 34: 307–311.
Nadal R, Zhang Z, Rahman H, Schweizer MT, Denmeade SR, Paller CJ et al. Clinical activity of enzalutamide in Docetaxel-naive and Docetaxel-pretreated patients with metastatic castration-resistant prostate cancer. Prostate 2014; 74: 1560–1568.
Hobisch A, Ramoner R, Fuchs D, Godoy-Tundidor S, Bartsch G, Klocker H et al. Prostate cancer cells (LNCaP) generated after long-term interleukin 6 (IL-6) treatment express IL-6 and acquire an IL-6 partially resistant phenotype. Clin Cancer Res 2001; 7: 2941–2948.
Hobisch A, Fritzer A, Comuzzi B, Fiechtl M, Malinowska K, Steiner H et al. The androgen receptor pathway is by-passed in prostate cancer cells generated after prolonged treatment with bicalutamide. Prostate 2006; 66: 413–420.
Puhr M, Hoefer J, Schafer G, Erb HH, Oh SJ, Klocker H et al. Epithelial-to-mesenchymal transition leads to docetaxel resistance in prostate cancer and is mediated by reduced expression of miR-200c and miR-205. Am J Pathol 2012; 181: 2188–2201.
Hoogland AM, Jenster G, van Weerden WM, Trapman J, van der Kwast T, Roobol MJ et al. ERG immunohistochemistry is not predictive for PSA recurrence, local recurrence or overall survival after radical prostatectomy for prostate cancer. Mod Pathol 2012; 25: 471–479.
Mikolcevic P, Sigl R, Rauch V, Hess MW, Pfaller K, Barisic M et al. Cyclin-dependent kinase 16/PCTAIRE kinase 1 is activated by cyclin Y and is essential for spermatogenesis. Mol Cell Biol 2012; 32: 868–879.
Puhr M, Santer FR, Neuwirt H, Susani M, Nemeth JA, Hobisch A et al. Down-regulation of suppressor of cytokine signaling-3 causes prostate cancer cell death through activation of the extrinsic and intrinsic apoptosis pathways. Cancer Res 2009; 69: 7375–7384.
Acknowledgements
We thank Irma Sottsas for TMA preparation, paraffin embedding and immunohistochemistry (IHC) stainings, Mag Eberhard Steiner for patient selection and statistical analysis, Dr Walter Parson for cell line authentication and Dr Yaron Galanty (University of Cambridge) for generously sharing PIAS1 constructs. This work was supported by the Austrian Cancer Society/Tirol, the intramural funding program of the Medical University of Innsbruck for young scientists MUI-START, Project 2010012007, Austrian Science Fund (FWF) Grants P 25639-B19 (to MP) and W1101-B18 (to ZC) and Anniversary Fund Austrian National Bank (ÖNB) Grant 15374 (to ZC).
Author contributions
MP established the project and postulated the hypothesis. He performed or supervised all experiments and wrote the manuscript together with JH. JH performed correlation and survival analyses, immunofluorescence stainings as well as R1881 treatments. AE performed luciferase assays and drug combination treatments. DD, BU, VS and GK constructed, stained and analyzed the Bonn TMA. GL and MH were responsible for the preparation of the Rotterdam TMA. FH cloned the Flag PIAS1 plasmid and helped to evaluate the AR ChIP-Seq data. BS analyzed together with MP the Innsbruck TMA. HN helped with patient statistics. HK was responsible for AR-ChIP-Seq data. ZC helped with the coordination of experiments. In addition, all co-authors helped improving the manuscript, and approved its final version.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Oncogene website
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Puhr, M., Hoefer, J., Eigentler, A. et al. PIAS1 is a determinant of poor survival and acts as a positive feedback regulator of AR signaling through enhanced AR stabilization in prostate cancer. Oncogene 35, 2322–2332 (2016). https://doi.org/10.1038/onc.2015.292
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2015.292
This article is cited by
-
Inhibition of CDKL3 downregulates STAT1 thus suppressing prostate cancer development
Cell Death & Disease (2023)
-
Quantitative SUMO proteomics identifies PIAS1 substrates involved in cell migration and motility
Nature Communications (2020)
-
Molecular Mechanisms of Enzalutamide Resistance in Prostate Cancer
Current Molecular Biology Reports (2017)