Abstract
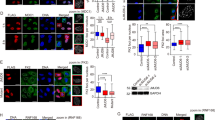
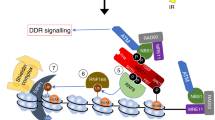
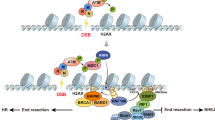
Chromatin remodeling factors are becoming known as crucial facilitators of recruitment of repair proteins to sites of DNA damage. Multiple chromatin remodeling protein complexes are now known to be required for efficient double strand break repair. In a screen for microRNAs (miRNAs) that modulate the DNA damage response, we discovered that expression of the miR-99 family of miRNAs correlates with radiation sensitivity. These miRNAs were also transiently induced following radiation. The miRNAs target the SWI/SNF chromatin remodeling factor SNF2H/SMARCA5, a component of the ACF1 complex. We found that by reducing levels of SNF2H, miR-99a and miR-100 reduced BRCA1 localization to sites of DNA damage. Introduction of the miR-99 family of miRNAs into cells reduced the rate and overall efficiency of repair by both homologous recombination and non-homologous end joining. Finally, induction of the miR-99 family following radiation prevents an increase in SNF2H expression and reduces the recruitment of BRCA1 to the sites of DNA damage following a second dose of radiation, reducing the efficiency of repair after multiple rounds of radiation, as used in fractionated radiotherapy.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
van Attikum H, Gasser SM . Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol 2009; 19: 207–217.
Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA et al. Genomic instability in mice lacking histone H2AX. Science 2002; 296: 922–927.
van Attikum H, Gasser SM . The histone code at DNA breaks: a guide to repair? Nat Rev Mol Cell Biol 2005; 6: 757–765.
van Attikum H, Fritsch O, Hohn B, Gasser SM . Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 2004; 119: 777–788.
van Attikum H, Fritsch O, Gasser SM . Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J 2007; 26: 4113–4125.
Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE et al. INO80 and [gamma]-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 2004; 119: 767–775.
Tsukuda T, Fleming AB, Nickoloff JA, Osley MA . Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 2005; 438: 379–383.
Lee H-S, Park J-H, Kim S-J, Kwon S-J, Kwon J . A cooperative activation loop among SWI/SNF, [gamma]-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J 2010; 29: 1434–1445.
Jha S, Shibata E, Dutta A . Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol Cell Biol 2008 28: 2690–2700.
Stewart GS, Wang B, Bignell CR, Taylor AMR, Elledge SJ . MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 2003; 421: 961–966.
Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 2007; 131: 887–900.
Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, FdrD Sweeney et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin Ligase. Science 2007; 318: 1637–1640.
Huen MSY, Grant R, Manke I, Minn K, Yu X, Yaffe MB et al. RNF8 Transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 2007; 131: 901–914.
Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 2009; 136: 435–446.
Roberts SA, Ramsden DA . Loading of the Nonhomologous End Joining Factor, Ku, on Protein-occluded DNA Ends. J Biol Chem 2007; 282: 10605–10613.
Mladenov E, Iliakis G . Induction and repair of DNA double strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutat Res Fundam Mol Mech Mugag 2011; 711: 61–72.
Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol 2010; 190: 731–740.
Lan L, Ui A, Nakajima S, Hatakeyama K, Hoshi M, Watanabe R et al. The ACF1 complex is required for DNA double-strand break repair in human cells. Mol Cell 2010; 40: 976–987.
Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DNP et al. Regulation of homologous recombination by rNF20-dependent H2B ubiquitination. Mol Cell 2011; 41: 515–528.
Sun D, Lee YS, Malhotra A, Kim HK, Matecic M, Evans C et al. miR-99 Family of MicroRNAs suppresses the expression of prostate-specific antigen and prostate cancer cell proliferation. Cancer Res 2011; 71: 1313–1324.
He L, Hannon GJ . MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004 5: 522–531.
Dey BK, Gagan J, Dutta A . miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol 2011; 31: 203–214.
Gagan J, Dey BK, Layer R, Yan Z, Dutta A . MICRORNA-378 targets the myogenic repressor myor during myoblast differentiation. J Biol Chem 2011; 22: 19432–19438.
Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY et al. Identification of a MicroRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol Cell 2011; 42: 500–510.
Chen C-Z, Li L, Lodish HF, Bartel DP . MicroRNAs modulate hematopoietic lineage differentiation. Science 2004; 303: 83–86.
Houbaviy HB, Murray MF, Sharp PA . Embryonic stem cell-specific MicroRNAs. Dev Cell 2003; 5: 351–358.
Lee YS, Dutta A . MicroRNAs in cancer. Annu Rev Pathol 2009; 4: 199–227.
Lee YS, Dutta A . The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev 2007; 21: 1025–1030.
He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S et al. A microRNA polycistron as a potential human oncogene. Nature 2005; 435: 828–833.
Chang T-C, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH et al. Transactivation of miR-34a by p53 broadly†influences gene expression and promotes apoptosis. Mol Cell 2007; 26: 745–752.
Marcu LG . Altered fractionation in radiotherapy: from radiobiological rationale to therapeutic gain. Cancer Treat Rev 2010; 36: 606–614.
Skvortsova I, Skvortsov S, Stasyk T, Raju U, Popper B-A, Schiestl B et al. Intracellular signaling pathways regulating radioresistance of human prostate carcinoma cells. Proteomics 2008; 8: 4521–4533.
Meng F, Henson R, Wehbe Ä, Janek H, Ghoshal K, Jacob ST et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007; 133: 647–658.
Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH et al. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev 2009; 23: 1327–1337.
Chan JA, Krichevsky AM, Kosik KS . MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 2005; 65: 6029–6033.
Sirotkin AV, Lauková M, Ovcharenko D, Brenaut P, Mlynček M . Identification of MicroRNAs controlling human ovarian cell proliferation and apoptosis. J Cell Physiol 2010; 223: 49–56.
Ransburgh DJR, Chiba N, Ishioka C, Toland AE, Parvin JD . Identification of breast tumor mutations in BRCA1 That abolish its function in homologous DNA recombination. Cancer Res 2010; 70: 988–995.
Pierce AJ, Johnson RD, Thompson LH, Jasin M . XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 1999; 13: 2633–2638.
Golding SE, Morgan RN, Adams BR, Hawkins AJ, Povirk LF, Valerie K et al. and ERK signaling from EGFR and mutant EGFRvIII enhances DNA double-strand break repair in human glioma cells. Cancer Biol Ther 2009; 8: 730–738.
Doghman M, Wakil AE, Cardinaud B, Thomas E, Wang J, Zhao W et al. Regulation of insulin-like growth factor,ÄìMammalian target of rapamycin signaling by MicroRNA in childhood adrenocortical tumors. Cancer Res 2010; 70: 4666–4675.
Hu H, Gatti RA . MicroRNAs: new players in the DNA damage response. J Mol Cell Biol 2011; 3: 151–158.
Lal A, Pan Y, Navarro F, Dykxhoorn DM, Moreau L, Meire E et al. miR-24-mediated downregulation of H2AX suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol 2009; 16: 492–498.
Wang P, Zou F, Zhang X, Li H, Dulak A, Tomko RJ et al. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res 2009; 69: 8157–8165.
He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y et al. A microRNA component of the p53 tumour suppressor network. Nature 2007; 447: 1130–1134.
Suzuki HI, Yamagata K, Sugimoto K, Iwamoto T, Kato S, Miyazono K . Modulation of microRNA processing by p53. Nature 2009; 460: 529–533.
Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell 2011; 41: 210–220.
Golding SE, Rosenberg E, Valerie N, Hussaini I, Frigerio M, Cockcroft XF et al. Improved ATM kinase inhibitor KU-60019 radiosensitizes glioma cells, compromises insulin, AKT and ERK prosurvival signaling, and inhibits migration and invasion. Mol Cancer Ther 2009; 8: 2894–2902.
Acknowledgements
AM was supported by DOD BCRP predoctoral traineeship BC073568. DS was supported by DOD PCRP predoctoral traineeship PC094499. This work was supported by P01CA104106 to AD. We thank members of the Dutta Laboratory for their help and discussion.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary information accompanies the paper on the Oncogene website
Rights and permissions
About this article
Cite this article
Mueller, A., Sun, D. & Dutta, A. The miR-99 family regulates the DNA damage response through its target SNF2H. Oncogene 32, 1164–1172 (2013). https://doi.org/10.1038/onc.2012.131
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2012.131
Keywords
This article is cited by
-
CircCASC15-miR-100-mTOR may influence the cervical cancer radioresistance
Cancer Cell International (2022)
-
MicroRNAs, DNA damage response and ageing
Biogerontology (2020)
-
Inhalation of lung spheroid cell secretome and exosomes promotes lung repair in pulmonary fibrosis
Nature Communications (2020)
-
Metformin overcomes resistance to cisplatin in triple-negative breast cancer (TNBC) cells by targeting RAD51
Breast Cancer Research (2019)
-
miR-125b-5p and miR-99a-5p downregulate human γδ T-cell activation and cytotoxicity
Cellular & Molecular Immunology (2019)