Abstract
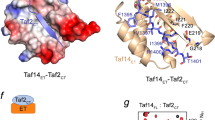
The general transcription factor TFIID provides a regulatory platform for transcription initiation. Here we present the crystal structure (1.97 Å) and NMR analysis of yeast TAF1 N-terminal domains TAND1 and TAND2 bound to yeast TBP, together with mutational data. We find that yeast TAF1-TAND1, which in itself acts as a transcriptional activator, binds TBP's concave DNA-binding surface by presenting similar anchor residues to TBP as does Mot1 but from a distinct structural scaffold. Furthermore, we show how TAF1-TAND2 uses an aromatic and acidic anchoring pattern to bind a conserved TBP surface groove traversing the basic helix region, and we find highly similar TBP-binding motifs also presented by the structurally distinct TFIIA, Mot1 and Brf1 proteins. Our identification of these anchoring patterns, which can be easily disrupted or enhanced, provides insight into the competitive multiprotein TBP interplay critical to transcriptional regulation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Goodrich, J.A. & Tjian, R. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat. Rev. Genet. 11, 549–558 (2010).
D'Alessio, J.A., Ng, R., Willenbring, H. & Tjian, R. Core promoter recognition complex changes accompany liver development. Proc. Natl. Acad. Sci. USA 108, 3906–3911 (2011).
Vannini, A. & Cramer, P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol. Cell 45, 439–446 (2012).
Papai, G., Weil, P.A. & Schultz, P. New insights into the function of transcription factor TFIID from recent structural studies. Curr. Opin. Genet. Dev. 21, 219–224 (2011).
Thomas, M.C. & Chiang, C.M. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41, 105–178 (2006).
Hahn, S. Structure and mechanism of the RNA polymerase II transcription machinery. Nat. Struct. Mol. Biol. 11, 394–403 (2004).
Bieniossek, C. et al. The architecture of human general transcription factor TFIID core complex. Nature 493, 699–702 (2013).
Chitikila, C., Huisinga, K.L., Irvin, J.D., Basehoar, A.D. & Pugh, B.F. Interplay of TBP inhibitors in global transcriptional control. Mol. Cell 10, 871–882 (2002).
Martel, L.S., Brown, H.J. & Berk, A.J. Evidence that TAF-TATA box-binding protein interactions are required for activated transcription in mammalian cells. Mol. Cell Biol. 22, 2788–2798 (2002).
Wollmann, P. et al. Structure and mechanism of the Swi2/Snf2 remodeller Mot1 in complex with its substrate TBP. Nature 475, 403–407 (2011).
Sigler, P.B. Transcriptional activation: acid blobs and negative noodles. Nature 333, 210–212 (1988).
Hahn, S. & Young, E.T. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 189, 705–736 (2011).
Ptashne, M. & Gann, A.A. Activators and targets. Nature 346, 329–331 (1990).
Brzovic, P.S. et al. The acidic transcription activator gcn4 binds the mediator subunit gal11/med15 using a simple protein interface forming a fuzzy complex. Mol. Cell 44, 942–953 (2011).
Liu, W.L. et al. Structures of three distinct activator-TFIID complexes. Genes Dev. 23, 1510–1521 (2009).
Baek, H.J., Kang, Y.K. & Roeder, R.G. Human Mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J. Biol. Chem. 281, 15172–15181 (2006).
Elmlund, H. et al. Cryo-EM reveals promoter DNA binding and conformational flexibility of the general transcription factor TFIID. Structure 17, 1442–1452 (2009).
Kotani, T. et al. A role of transcriptional activators as antirepressors for the autoinhibitory activity of TATA box binding of transcription factor IID. Proc. Natl. Acad. Sci. USA 97, 7178–7183 (2000).
Muldrow, T.A., Campbell, A.M., Weil, P.A. & Auble, D.T. MOT1 can activate basal transcription in vitro by regulating the distribution of TATA binding protein between promoter and nonpromoter sites. Mol. Cell Biol. 19, 2835–2845 (1999).
Mal, T.K. et al. Structural and functional characterization on the interaction of yeast TFIID subunit TAF1 with TATA-binding protein. J. Mol. Biol. 339, 681–693 (2004).
Takahata, S., Kasahara, K., Kawaichi, M. & Kokubo, T. Autonomous function of the amino-terminal inhibitory domain of TAF1 in transcriptional regulation. Mol. Cell Biol. 24, 3089–3099 (2004).
Bagby, S. et al. TFIIA-TAF regulatory interplay: NMR evidence for overlapping binding sites on TBP. FEBS Lett. 468, 149–154 (2000).
Kokubo, T., Swanson, M.J., Nishikawa, J.I., Hinnebusch, A.G. & Nakatani, Y. The yeast TAF145 inhibitory domain and TFIIA competitively bind to TATA-binding protein. Mol. Cell Biol. 18, 1003–1012 (1998).
Liu, D. et al. Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell 94, 573–583 (1998).
Mal, T.K. et al. Functional silencing of TATA-binding protein (TBP) by a covalent linkage of the N-terminal domain of TBP-associated factor 1. J. Biol. Chem. 282, 22228–22238 (2007).
Kim, J.L., Nikolov, D.B. & Burley, S.K. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature 365, 520–527 (1993).
Buratowski, S. & Zhou, H. Transcription factor IID mutants defective for interaction with transcription factor IIA. Science 255, 1130–1132 (1992).
Kim, T.K. & Roeder, R.G. Involvement of the basic repeat domain of TATA-binding protein (TBP) in transcription by RNA polymerases I, II, and III. J. Biol. Chem. 269, 4891–4894 (1994).
Kotani, T. et al. Identification of highly conserved amino-terminal segments of dTAFII230 and yTAFII145 that are functionally interchangeable for inhibiting TBP-DNA interactions in vitro and in promoting yeast cell growth in vivo. J. Biol. Chem. 273, 32254–32264 (1998).
Ozer, J., Mitsouras, K., Zerby, D., Carey, M. & Lieberman, P.M. Transcription factor IIA derepresses TATA-binding protein (TBP)-associated factor inhibition of TBP-DNA binding. J. Biol. Chem. 273, 14293–14300 (1998).
Juo, Z.S., Kassavetis, G.A., Wang, J., Geiduschek, E.P. & Sigler, P.B. Crystal structure of a transcription factor IIIB core interface ternary complex. Nature 422, 534–539 (2003).
Lee, D.K., DeJong, J., Hashimoto, S., Horikoshi, M. & Roeder, R.G. TFIIA induces conformational changes in TFIID via interactions with the basic repeat. Mol. Cell Biol. 12, 5189–5196 (1992).
Solow, S.P., Lezina, L. & Lieberman, P.M. Phosphorylation of TFIIA stimulates TATA binding protein-TATA interaction and contributes to maximal transcription and viability in yeast. Mol. Cell Biol. 19, 2846–2852 (1999).
Solow, S., Salunek, M., Ryan, R. & Lieberman, P.M. Taf(II) 250 phosphorylates human transcription factor IIA on serine residues important for TBP binding and transcription activity. J. Biol. Chem. 276, 15886–15892 (2001).
Bleichenbacher, M., Tan, S. & Richmond, T.J. Novel interactions between the components of human and yeast TFIIA/TBP/DNA complexes. J. Mol. Biol. 332, 783–793 (2003).
Woiwode, A. et al. PTEN represses RNA polymerase III-dependent transcription by targeting the TFIIIB complex. Mol. Cell Biol. 28, 4204–4214 (2008).
Garza, A.M., Khan, S.H. & Kumar, R. Site-specific phosphorylation induces functionally active conformation in the intrinsically disordered N-terminal activation function (AF1) domain of the glucocorticoid receptor. Mol. Cell Biol. 30, 220–230 (2010).
Wright, P.E. & Dyson, H.J. Linking folding and binding. Curr. Opin. Struct. Biol. 19, 31–38 (2009).
Hoeflich, K.P. & Ikura, M. Calmodulin in action: diversity in target recognition and activation mechanisms. Cell 108, 739–742 (2002).
Lee, C.W., Martinez-Yamout, M.A., Dyson, H.J. & Wright, P.E. Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding protein. Biochemistry 49, 9964–9971 (2010).
van Werven, F.J., van Teeffelen, H.A., Holstege, F.C. & Timmers, H.T. Distinct promoter dynamics of the basal transcription factor TBP across the yeast genome. Nat. Struct. Mol. Biol. 16, 1043–1048 (2009).
Liu, J. et al. Intrinsic disorder in transcription factors. Biochemistry 45, 6873–6888 (2006).
Kim, Y., Geiger, J.H., Hahn, S. & Sigler, P.B. Crystal structure of a yeast TBP/TATA-box complex. Nature 365, 512–520 (1993).
Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Blanc, E. et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr. D Biol. Crystallogr. 60, 2210–2221 (2004).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Willard, L. et al. VADAR: a web server for quantitative evaluation of protein structure quality. Nucleic Acids Res. 31, 3316–3319 (2003).
Andresen, C. et al. Transient structure and dynamics in the disordered c-Myc transactivation domain affect Bin1 binding. Nucleic Acids Res. 40, 6353–6366 (2012).
Mal, T.K. et al. Resonance assignments of 30 kDa complexes of TFIID subunit TAF1 with TATA-binding protein. J. Biomol. NMR 33, 76 (2005).
Ahlner, A., Carlsson, M., Jonsson, B.H. & Lundström, P. PINT: a software for integration of peak volumes and extraction of relaxation rates. J. Biomol. NMR published online, http://dx.doi.org/10.1007/s10858-013-9737-7 (9 May 2013).
Mosteller, F. & Tukey, J.W. Data Analysis and Regression: A Second Course in Statistics 133–162 (Addison-Wesley, 1977).
Palmer, A.G. III & Massi, F. Characterization of the dynamics of biomacromolecules using rotating-frame spin relaxation NMR spectroscopy. Chem. Rev. 106, 1700–1719 (2006).
Dosset, P., Hus, J.C., Blackledge, M. & Marion, D. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J. Biomol. NMR 16, 23–28 (2000).
Ohyama, Y., Kasahara, K. & Kokubo, T. Saccharomyces cerevisiae Ssd1p promotes CLN2 expression by binding to the 5′-untranslated region of CLN2 mRNA. Genes Cells 15, 1169–1188 (2010).
Takahashi, H., Kasahara, K. & Kokubo, T. Saccharomyces cerevisiae Med9 comprises two functionally distinct domains that play different roles in transcriptional regulation. Genes Cells 14, 53–67 (2009).
Kunkel, T.A., Roberts, J.D. & Zakour, R.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154, 367–382 (1987).
Sikorski, R.S. & Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 (1989).
Takahata, S. et al. Identification of a novel TATA element-binding protein binding region at the N terminus of the Saccharomyces cerevisiae TAF1 protein. J. Biol. Chem. 278, 45888–45902 (2003).
Acknowledgements
This work was supported by the Swedish Research Council (621-2011-6028 and 621-2012-5250 to M.S.; 621-2012-5136 to P.L.), VINNOVA (P32045-1 to M.S.), the Swedish Cancer Foundation (11 0681 to M.S.), the Swedish Child Cancer Foundation (PROJ09/092 to M.S.), the Forum Scientium Award (C.A.), the Canadian Institutes for Health Research (MT-13611 to M.I.), the grant-in-Aid for Scientific Research from Japan Society for the Promotion of Science (23370077 to T.K.) and equipment grants to Linköping University from the Knut and Alice Wallenberg foundation. M.I. is supported as a Canada Research Chair. We thank H.Th.M. Timmers, L. Penn, C.H. Arrowsmith and P. de Graaf for critical discussion and acknowledge the Swedish NMR Centre, the Protein Science Facility and beamline ID14-1 at the European Synchrotron Radiation Facility.
Author information
Authors and Affiliations
Contributions
M.I. and M.S. conceived of and designed the study. Experiments and data evaluation were designed and performed by M.A., M.I.S. and M.M. (crystallography), Y.O. and T.K. (yeast mutations) and M.A., C.A., S.H., P.L. and M.S. (NMR). M.A., C.A., S.H., M.M. and M.S. wrote the paper. All authors discussed the interpretation and implications of the results and edited the manuscript at all stages.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figures and Tables
Supplementary Figures 1–4 and Supplementary Tables 1–3 (PDF 3035 kb)
Rights and permissions
About this article
Cite this article
Anandapadamanaban, M., Andresen, C., Helander, S. et al. High-resolution structure of TBP with TAF1 reveals anchoring patterns in transcriptional regulation. Nat Struct Mol Biol 20, 1008–1014 (2013). https://doi.org/10.1038/nsmb.2611
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2611
This article is cited by
-
Structural convergence endows nuclear transport receptor Kap114p with a transcriptional repressor function toward TATA-binding protein
Nature Communications (2023)
-
H3K4me2/3 modulate the stability of RNA polymerase II pausing
Cell Research (2023)
-
Ino2, activator of yeast phospholipid biosynthetic genes, interacts with basal transcription factors TFIIA and Bdf1
Current Genetics (2023)
-
Trimeric complexes of Antp-TBP with TFIIEβ or Exd modulate transcriptional activity
Hereditas (2022)
-
NAUTICA: classifying transcription factor interactions by positional and protein-protein interaction information
Biology Direct (2020)