Key Points
-
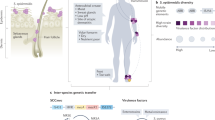
Staphylococcus epidermidis is a common member of the human epithelial microflora and one of the most frequent nosocomial pathogens.
-
S. epidermidis is mostly involved with indwelling medical device-associated infections.The prevalence of S. epidermidis in this type of infection is likely to be due to its abundance on the human skin and its capacity to adhere to catheter surfaces and form biofilms.
-
Biofilm formation, exopolymers and other mechanisms protect S. epidermidis from antibiotics and host defences.
-
Efficient S. epidermidis biofilm formation is dependent on both protein and exopolysaccharide aggregation substances.
-
S. epidermidis can sense the presence of antimicrobial peptides and trigger defensive responses against this type of innate host defence mechanism, which it encounters in its natural habitat.
-
S. epidermidis functions as a reservoir for genes that can be transferred to Staphylococcus aureus, enhancing the pathogenic success and antibiotic resistance of this more dangerous pathogen.
-
S. epidermidis does not produce aggressive toxins and its immune evasion factors probably have original functions in the commensal lifestyle of this species. This indicates that S. epidermidis infection is 'accidental' in nature.
Abstract
Although nosocomial infections by Staphylococcus epidermidis have gained much attention, this skin-colonizing bacterium has apparently evolved not to cause disease, but to maintain the commonly benign relationship with its host. Accordingly, S. epidermidis does not produce aggressive virulence determinants. Rather, factors that normally sustain the commensal lifestyle of S. epidermidis seem to give rise to additional benefits during infection. Furthermore, we are beginning to comprehend the roles of S. epidermidis in balancing the epithelial microflora and serving as a reservoir of resistance genes. In this Review, I discuss the molecular basis of the commensal and infectious lifestyles of S. epidermidis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
CDC. National Nosocomial Infections Surveillance (NNIS) system report, data summary from January 1992 through June 2004, issued October 2004. Am. J. Infect. Control 32, 470–485 (2004).
Uckay, I. et al. Foreign body infections due to Staphylococcus epidermidis. Ann. Med. 41, 109–119 (2009).
Dimick, J. B. et al. Increased resource use associated with catheter-related bloodstream infection in the surgical intensive care unit. Arch. Surg. 136, 229–234 (2001).
Rello, J. et al. Evaluation of outcome of intravenous catheter-related infections in critically ill patients. Am. J. Respir. Crit. Care Med. 162, 1027–1030 (2000).
Rogers, K. L., Fey, P. D. & Rupp, M. E. Coagulase-negative staphylococcal infections. Infect. Dis. Clin. North Am. 23, 73–98 (2009). This provides an excellent review on clinical aspects of S. epidermidis infections.
Costerton, J. W., Stewart, P. S. & Greenberg, E. P. Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322 (1999).
Kloos, W. & Schleifer, K. H. in Bergey's Manual of Systematic Bacteriology (eds Sneath, P. H. A., Mair, N., Sharpe, M. E. & Holt, J. G.) 1013–1035 (Williams & Wilkins, Baltimore, 1986).
Kloos, W. E. & Musselwhite, M. S. Distribution and persistence of Staphylococcus and Micrococcus species and other aerobic bacteria on human skin. Appl. Microbiol. 30, 381–385 (1975).
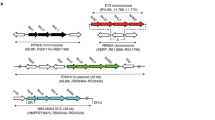
Gill, S. R. et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187, 2426–2438 (2005). This article describes the sequencing and comparison of the genomes of biofilm-forming S. epidermidis and S. aureus.
Zhang, Y. Q. et al. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 49, 1577–1593 (2003).
Wang, X. M. et al. Evaluation of a multilocus sequence typing system for Staphylococcus epidermidis. J. Med. Microbiol. 52, 989–998 (2003).
Wisplinghoff, H. et al. Related clones containing SCCmec type IV predominate among clinically significant Staphylococcus epidermidis isolates. Antimicrob. Agents Chemother. 47, 3574–3579 (2003).
Thomas, J. C. et al. Improved multilocus sequence typing scheme for Staphylococcus epidermidis. J. Clin. Microbiol. 45, 616–619 (2007).
Miragaia, M., Thomas, J. C., Couto, I., Enright, M. C. & de Lencastre, H. Inferring a population structure for Staphylococcus epidermidis from multilocus sequence typing data. J. Bacteriol. 189, 2540–2552 (2007). This article details an investigation of the population structure of S. epidermidis.
Li, M., Wang, X., Gao, Q. & Lu, Y. Molecular characterization of Staphylococcus epidermidis strains isolated from a teaching hospital in Shanghai, China. J. Med. Microbiol. 58, 456–461 (2009).
Galdbart, J. O., Allignet, J., Tung, H. S., Ryden, C. & El Solh, N. Screening for Staphylococcus epidermidis markers discriminating between skin-flora strains and those responsible for infections of joint prostheses. J. Infect. Dis. 182, 351–355 (2000).
Gu, J. et al. Bacterial insertion sequence IS256 as a potential molecular marker to discriminate invasive strains from commensal strains of Staphylococcus epidermidis. J. Hosp. Infect. 61, 342–348 (2005).
Kozitskaya, S. et al. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect. Immun. 72, 1210–1215 (2004).
Yao, Y. et al. Factors characterizing Staphylococcus epidermidis invasiveness determined by comparative genomics. Infect. Immun. 73, 1856–1860 (2005).
Lina, G. et al. Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl. Environ. Microbiol. 69, 18–23 (2003).
O'Grady, N. P. et al. Guidelines for the prevention of intravascular catheter-related infections. MMWR Recomm. Rep. 51, 1–26 (2002).
Chu, V. H. et al. Coagulase-negative staphylococcal prosthetic valve endocarditis — a contemporary update based on the International Collaboration on Endocarditis: prospective cohort study. Heart 95, 570–576 (2009).
Massey, R. C., Horsburgh, M. J., Lina, G., Hook, M. & Recker, M. The evolution and maintenance of virulence in Staphylococcus aureus: a role for host-to-host transmission? Nature Rev. Microbiol. 4, 953–958 (2006). This article reviews the mathematical model that explains the evolution of lifestyle differences between S. epidermidis and S. aureus.
Otto, M., Sussmuth, R., Vuong, C., Jung, G. & Gotz, F. Inhibition of virulence factor expression in Staphylococcus aureus by the Staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett. 450, 257–262 (1999).
Carmody, A. B. & Otto, M. Specificity grouping of the accessory gene regulator quorum-sensing system of Staphylococcus epidermidis is linked to infection. Arch. Microbiol. 181, 250–253 (2004).
Harder, J. & Schroder, J. M. Antimicrobial peptides in human skin. Chem. Immunol. Allergy 86, 22–41 (2005).
Faurschou, M. & Borregaard, N. Neutrophil granules and secretory vesicles in inflammation. Microbes Infect. 5, 1317–1327 (2003).
Pourmand, M. R., Clarke, S. R., Schuman, R. F., Mond, J. J. & Foster, S. J. Identification of antigenic components of Staphylococcus epidermidis expressed during human infection. Infect. Immun. 74, 4644–4654 (2006).
Yao, Y., Sturdevant, D. E. & Otto, M. Genomewide analysis of gene expression in Staphylococcus epidermidis biofilms: insights into the pathophysiology of S. epidermidis biofilms and the role of phenol-soluble modulins in formation of biofilms. J. Infect. Dis. 191, 289–298 (2005). An investigation of genome-wide gene regulatory changes that occur in S. epidermidis biofilms.
Khardori, N., Yassien, M. & Wilson, K. Tolerance of Staphylococcus epidermidis grown from indwelling vascular catheters to antimicrobial agents. J. Ind. Microbiol. 15, 148–151 (1995).
Duguid, I. G., Evans, E., Brown, M. R. & Gilbert, P. Effect of biofilm culture upon the susceptibility of Staphylococcus epidermidis to tobramycin. J. Antimicrobiol. Chemother. 30, 803–810 (1992).
Duguid, I. G., Evans, E., Brown, M. R. & Gilbert, P. Growth-rate-independent killing by ciprofloxacin of biofilm-derived Staphylococcus epidermidis; evidence for cell-cycle dependency. J. Antimicrobiol. Chemother. 30, 791–802 (1992).
O'Toole, G., Kaplan, H. B. & Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54, 49–79 (2000).
Vacheethasanee, K. et al. Bacterial surface properties of clinically isolated Staphylococcus epidermidis strains determine adhesion on polyethylene. J. Biomed. Mater. Res. 42, 425–432 (1998).
Heilmann, C., Hussain, M., Peters, G. & Gotz, F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24, 1013–1024 (1997).
Tormo, M. A., Knecht, E., Gotz, F., Lasa, I. & Penades, J. R. Bap-dependent biofilm formation by pathogenic species of Staphylococcus: evidence of horizontal gene transfer? Microbiology 151, 2465–2475 (2005).
Mazmanian, S. K., Liu, G., Ton-That, H. & Schneewind, O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285, 760–763 (1999).
Navarre, W. W. & Schneewind, O. Surface proteins of Gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63, 174–229 (1999).
Arrecubieta, C., Lee, M. H., Macey, A., Foster, T. J. & Lowy, F. D. SdrF, a Staphylococcus epidermidis surface protein, binds type I collagen. J. Biol. Chem. 282, 18767–18776 (2007).
Hartford, O., O'Brien, L., Schofield, K., Wells, J. & Foster, T. J. The Fbe (SdrG) protein of Staphylococcus epidermidis HB promotes bacterial adherence to fibrinogen. Microbiology 147, 2545–2552 (2001).
Heilmann, C. et al. Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology 149, 2769–2778 (2003).
McCrea, K. W. et al. The serine-aspartate repeat (Sdr) protein family in Staphylococcus epidermidis. Microbiology 146, 1535–1546 (2000).
Nilsson, M. et al. A fibrinogen-binding protein of Staphylococcus epidermidis. Infect. Immun. 66, 2666–2673 (1998).
Guo, B., Zhao, X., Shi, Y., Zhu, D. & Zhang, Y. Pathogenic implication of a fibrinogen-binding protein of Staphylococcus epidermidis in a rat model of intravascular-catheter-associated infection. Infect. Immun. 75, 2991–2995 (2007).
Ponnuraj, K. et al. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell 115, 217–228 (2003). This work elucidated the mechanism by which SdrG binds to fibrinogen.
Sellman, B. R. et al. Expression of Staphylococcus epidermidis SdrG increases following exposure to an in vivo environment. Infect. Immun. 76, 2950–2957 (2008).
Arrecubieta, C. et al. SdrF, a Staphylococcus epidermidis surface protein, contributes to the initiation of ventricular assist device driveline-related infections. PLoS Pathog. 5, e1000411 (2009).
Bowden, M. G. et al. Identification and preliminary characterization of cell-wall-anchored proteins of Staphylococcus epidermidis. Microbiology 151, 1453–1464 (2005).
Gross, M., Cramton, S. E., Gotz, F. & Peschel, A. Key role of teichoic acid net charge in Staphylococcus aureus colonization of artificial surfaces. Infect. Immun. 69, 3423–3426 (2001).
Sadovskaya, I., Vinogradov, E., Flahaut, S., Kogan, G. & Jabbouri, S. Extracellular carbohydrate-containing polymers of a model biofilm-producing strain, Staphylococcus epidermidis RP62A. Infect. Immun. 73, 3007–3017 (2005).
Rice, K. C. et al. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl Acad. Sci. USA 104, 8113–8118 (2007).
Mack, D. et al. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178, 175–183 (1996). This article details the structural characterization of the exopolysaccharide PNAG.
Darby, C., Hsu, J. W., Ghori, N. & Falkow, S. Caenorhabditis elegans: plague bacteria biofilm blocks food intake. Nature 417, 243–244 (2002).
Wang, X., Preston, J. F. I. & Romeo, T. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186, 2724–2734 (2004).
Heilmann, C., Gerke, C., Perdreau-Remington, F. & Gotz, F. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect. Immun. 64, 277–282 (1996).
Heilmann, C. et al. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20, 1083–1091 (1996). This work identified the genetic locus that governs PNAG biosynthesis.
Francois, P. et al. Lack of biofilm contribution to bacterial colonisation in an experimental model of foreign body infection by Staphylococcus aureus and Staphylococcus epidermidis. FEMS Immunol. Med. Microbiol. 35, 135–140 (2003).
Rupp, M. E., Ulphani, J. S., Fey, P. D., Bartscht, K. & Mack, D. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect. Immun. 67, 2627–2632 (1999). This was the first in vivo analysis of an S. epidermidis virulence determinant (PNAG) using an isogenic mutant.
Rupp, M. E., Ulphani, J. S., Fey, P. D. & Mack, D. Characterization of Staphylococcus epidermidis polysaccharide intercellular adhesin/hemagglutinin in the pathogenesis of intravascular catheter-associated infection in a rat model. Infect. Immun. 67, 2656–2659 (1999).
Chokr, A., Leterme, D., Watier, D. & Jabbouri, S. Neither the presence of ica locus, nor in vitro-biofilm formation ability is a crucial parameter for some Staphylococcus epidermidis strains to maintain an infection in a guinea pig tissue cage model. Microb. Pathog. 42, 94–97 (2007).
Fluckiger, U. et al. Biofilm formation, icaADBC transcription, and polysaccharide intercellular adhesin synthesis by staphylococci in a device-related infection model. Infect. Immun. 73, 1811–1819 (2005).
Gerke, C., Kraft, A., Sussmuth, R., Schweitzer, O. & Gotz, F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 273, 18586–18593 (1998). This article describes the identification of the biochemical function of the IcaA and IcaD proteins.
Vuong, C. et al. A crucial role for exopolysaccharide modification in bacterial biofilm formation, immune evasion, and virulence. J. Biol. Chem. 279, 54881–54886 (2004). This study identified the biochemical function of the IcaB PNAG de-acetylase and showed its role in vitro and in vivo.
O'Gara, J. P. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol. Lett. 270, 179–188 (2007).
Ziebuhr, W. et al. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32, 345–356 (1999).
Knobloch, J. K. et al. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183, 2624–2633 (2001).
Mack, D. et al. Identification of three essential regulatory gene loci governing expression of Staphylococcus epidermidis polysaccharide intercellular adhesin and biofilm formation. Infect. Immun. 68, 3799–3807 (2000).
Tormo, M. A. et al. SarA is an essential positive regulator of Staphylococcus epidermidis biofilm development. J. Bacteriol. 187, 2348–2356 (2005).
Handke, L. D. et al. σB and SarA independently regulate polysaccharide intercellular adhesin production in Staphylococcus epidermidis. Can. J. Microbiol. 53, 82–91 (2007).
Al Laham, N. et al. Augmented expression of polysaccharide intercellular adhesin in a defined Staphylococcus epidermidis mutant with the small-colony-variant phenotype. J. Bacteriol. 189, 4494–4501 (2007).
Xu, L. et al. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect. Immun. 74, 488–496 (2006).
Vuong, C., Gerke, C., Somerville, G. A., Fischer, E. R. & Otto, M. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J. Infect. Dis. 188, 706–718 (2003).
Kogan, G., Sadovskaya, I., Chaignon, P., Chokr, A. & Jabbouri, S. Biofilms of clinical strains of Staphylococcus that do not contain polysaccharide intercellular adhesin. FEMS Microbiol. Lett. 255, 11–16 (2006).
Rohde, H. et al. Polysaccharide intercellular adhesin or protein factors in biofilm accumulation of Staphylococcus epidermidis and Staphylococcus aureus isolated from prosthetic hip and knee joint infections. Biomaterials 28, 1711–1720 (2007). This article gives an exceptionally balanced view of the roles of proteins versus exopolysaccharide in S. epidermis biofilm formation, in contrast to several reports that point to the importance of protein-mediated biofilm formation.
Hussain, M., Herrmann, M., von Eiff, C., Perdreau-Remington, F. & Peters, G. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect. Immun. 65, 519–524 (1997).
Rohde, H. et al. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 55, 1883–1895 (2005).
Conrady, D. G. et al. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc. Natl Acad. Sci. USA 105, 19456–19461 (2008). This work shed light on the mechanism of Aap self-aggregation.
Banner, M. A. et al. Localized tufts of fibrils on Staphylococcus epidermidis NCTC 11047 are comprised of the accumulation-associated protein. J. Bacteriol. 189, 2793–2804 (2007).
Bateman, A., Holden, M. T. & Yeats, C. The G5 domain: a potential N-acetylglucosamine recognition domain involved in biofilm formation. Bioinformatics 21, 1301–1303 (2005).
Sun, D., Accavitti, M. A. & Bryers, J. D. Inhibition of biofilm formation by monoclonal antibodies against Staphylococcus epidermidis RP62A accumulation-associated protein. Clin. Diagn. Lab. Immunol. 12, 93–100 (2005).
Conlon, K. M., Humphreys, H. & O'Gara, J. P. Inactivations of rsbU and sarA by IS256 represent novel mechanisms of biofilm phenotypic variation in Staphylococcus epidermidis. J. Bacteriol. 186, 6208–6219 (2004).
Chaignon, P. et al. Susceptibility of staphylococcal biofilms to enzymatic treatments depends on their chemical composition. Appl. Microbiol. Biotechnol. 75, 125–132 (2007).
Vuong, C., Kocianova, S., Yao, Y., Carmody, A. B. & Otto, M. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J. Infect. Dis. 190, 1498–1505 (2004). This manuscript shows the role of the S. epidermidis agr quorum sensing regulator in vivo.
Yarwood, J. M., Bartels, D. J., Volper, E. M. & Greenberg, E. P. Quorum sensing in Staphylococcus aureus biofilms. J. Bacteriol. 186, 1838–1850 (2004).
Boles, B. R. & Horswill, A. R. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 4, e1000052 (2008).
Teufel, P. & Gotz, F. Characterization of an extracellular metalloprotease with elastase activity from Staphylococcus epidermidis. J. Bacteriol. 175, 4218–4224 (1993).
Dubin, G. et al. Molecular cloning and biochemical characterisation of proteases from Staphylococcus epidermidis. Biol. Chem. 382, 1575–1582 (2001).
Ohara-Nemoto, Y. et al. Characterization and molecular cloning of a glutamyl endopeptidase from Staphylococcus epidermidis. Microb. Pathog. 33, 33–41 (2002).
Kaplan, J. B. et al. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J. Bacteriol. 186, 8213–8220 (2004).
Kaplan, J. B., Ragunath, C., Ramasubbu, N. & Fine, D. H. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous β-hexosaminidase activity. J. Bacteriol. 185, 4693–4698 (2003).
Kong, K. F., Vuong, C. & Otto, M. Staphylococcus quorum sensing in biofilm formation and infection. Int. J. Med. Microbiol. 296, 133–139 (2006).
Vuong, C. et al. Regulated expression of pathogen-associated molecular pattern molecules in Staphylococcus epidermidis: quorum-sensing determines pro-inflammatory capacity and production of phenol-soluble modulins. Cell. Microbiol. 6, 753–759 (2004).
Yao, Y. et al. Characterization of the Staphylococcus epidermidis accessory-gene regulator response: quorum-sensing regulation of resistance to human innate host defence. J. Infect. Dis. 193, 841–848 (2006).
Kocianova, S. et al. Key role of poly-γ-DL-glutamic acid in immune evasion and virulence of Staphylococcus epidermidis. J. Clin. Invest. 115, 688–694 (2005). This article investigates the role of poly-γ- DL -glutamic acid in S. epidermidis.
Little, S. F. & Ivins, B. E. Molecular pathogenesis of Bacillus anthracis infection. Microbes Infect. 1, 131–139 (1999).
Oppermann-Sanio, F. B. & Steinbuchel, A. Occurrence, functions and biosynthesis of polyamides in microorganisms and biotechnological production. Naturwissenschaften 89, 11–22 (2002).
Kristian, S. A. et al. Biofilm formation induces C3a release and protects Staphylococcus epidermidis from IgG and complement deposition and from neutrophil-dependent killing. J. Infect. Dis. 197, 1028–1035 (2008).
Vuong, C. et al. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell. Microbiol. 6, 269–275 (2004). This study shows the important role of PNAG in immune evasion.
Begun, J. et al. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defences. PLoS Pathog. 3, e57 (2007).
Mah, T. F. et al. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature 426, 306–310 (2003).
Heine, H. & Ulmer, A. J. Recognition of bacterial products by Toll-like receptors. Chem. Immunol. Allergy 86, 99–119 (2005).
Stevens, N. T. et al. Staphylococcus epidermidis polysaccharide intercellular adhesin induces IL-8 expression in human astrocytes via a mechanism involving TLR2. Cell. Microbiol. 11, 421–432 (2008).
Henneke, P. et al. Lipoproteins are critical TLR2 activating toxins in group B streptococcal sepsis. J. Immunol. 180, 6149–6158 (2008).
Li, H., Nooh, M. M., Kotb, M. & Re, F. Commercial peptidoglycan preparations are contaminated with superantigen-like activity that stimulates IL-17 production. J. Leukoc. Biol. 83, 409–418 (2008).
Hashimoto, M. et al. Not lipoteichoic acid but lipoproteins appear to be the dominant immunobiologically active compounds in Staphylococcus aureus. J. Immunol. 177, 3162–3169 (2006).
Mehlin, C., Headley, C. M. & Klebanoff, S. J. An inflammatory polypeptide complex from Staphylococcus epidermidis: isolation and characterization. J. Exp. Med. 189, 907–918 (1999). This article describes the identification and pro-inflammatory properties of the main S. epidermidis PSMs.
Wang, R. et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nature Med. 13, 1510–1514 (2007).
Hajjar, A. M. et al. Cutting edge: functional interactions between Toll-like receptor (TLR) 2 and TLR1 or TLR6 in response to phenol-soluble modulin. J. Immunol. 166, 15–19 (2001).
Lambert, P. A., Worthington, T., Tebbs, S. E. & Elliott, T. S. Lipid S, a novel Staphylococcus epidermidis exocellular antigen with potential for the serodiagnosis of infections. FEMS Immunol. Med. Microbiol. 29, 195–202 (2000).
Li, M. et al. Gram-positive three-component antimicrobial peptide-sensing system. Proc. Natl Acad. Sci. USA 104, 9469–9474 (2007). This work identified and characterized the first Gram-positive AMP sensor in S. epidermidis.
Peschel, A. et al. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274, 8405–8410 (1999).
Peschel, A. et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with L-lysine. J. Exp. Med. 193, 1067–1076 (2001).
Li, M. et al. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol. Microbiol. 66, 1136–1147 (2007).
Bader, M. W. et al. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122, 461–472 (2005).
Marin, M. E., de la Rosa, M. C. & Cornejo, I. Enterotoxigenicity of Staphylococcus strains isolated from Spanish dry-cured hams. Appl. Environ. Microbiol. 58, 1067–1069 (1992).
Bautista, L., Gaya, P., Medina, M. & Nunez, M. A quantitative study of enterotoxin production by sheep milk staphylococci. Appl. Environ. Microbiol. 54, 566–569 (1988).
Klingenberg, C. et al. Persistent strains of coagulase-negative staphylococci in a neonatal intensive care unit: virulence factors and invasiveness. Clin. Microbiol. Infect. 13, 1100–1111 (2007).
Scheifele, D. W., Bjornson, G. L., Dyer, R. A. & Dimmick, J. E. Delta-like toxin produced by coagulase-negative staphylococci is associated with neonatal necrotizing enterocolitis. Infect. Immun. 55, 2268–2273 (1987).
Rohde, H. et al. Detection of virulence-associated genes not useful for discriminating between invasive and commensal Staphylococcus epidermidis strains from a bone marrow transplant unit. J. Clin. Microbiol. 42, 5614–5619 (2004).
Ziebuhr, W. et al. Modulation of the polysaccharide intercellular adhesin (PIA) expression in biofilm forming Staphylococcus epidermidis. Analysis of genetic mechanisms. Adv. Exp. Med. Biol. 485, 151–157 (2000).
Rogers, K. L., Rupp, M. E. & Fey, P. D. The presence of icaADBC is detrimental to the colonization of human skin by Staphylococcus epidermidis. Appl. Environ. Microbiol. 74, 6155–6157 (2008).
Lai, Y. et al. The human anionic antimicrobial peptide dermcidin induces proteolytic defence mechanisms in staphylococci. Mol. Microbiol. 63, 497–506 (2007).
Diekema, D. J. et al. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997–1999. Clin. Infect. Dis. 32, S114–S132 (2001).
Vos, M. C., Ott, A. & Verbrugh, H. A. Successful search-and-destroy policy for methicillin-resistant Staphylococcus aureus in The Netherlands. J. Clin. Microbiol. 43, 2034; author reply 2034–2035 (2005).
van Pelt, C. et al. Strict infection control measures do not prevent clonal spread of coagulase negative staphylococci colonizing central venous catheters in neutropenic hemato-oncologic patients. FEMS Immunol. Med. Microbiol. 38, 153–158 (2003).
Chambers, H. F., Hartman, B. J. & Tomasz, A. Increased amounts of a novel penicillin-binding protein in a strain of methicillin-resistant Staphylococcus aureus exposed to nafcillin. J. Clin. Invest. 76, 325–331 (1985).
Ma, X. X. et al. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46, 1147–1152 (2002).
Miragaia, M., Couto, I. & de Lencastre, H. Genetic diversity among methicillin-resistant Staphylococcus epidermidis (MRSE). Microb. Drug Resist. 11, 83–93 (2005).
Diep, B. A. et al. The arginine catabolic mobile element and staphylococcal chromosomal cassette mec linkage: convergence of virulence and resistance in the USA300 clone of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 197, 1523–1530 (2008).
Miragaia, M. et al. Molecular characterization of methicillin-resistant Staphylococcus epidermidis clones: evidence of geographic dissemination. J. Clin. Microbiol. 40, 430–438 (2002).
Raad, I., Hanna, H. & Maki, D. Intravascular catheter-related infections: advances in diagnosis, prevention, and management. Lancet Infect. Dis. 7, 645–657 (2007).
Schwalbe, R. S., Stapleton, J. T. & Gilligan, P. H. Emergence of vancomycin resistance in coagulase-negative staphylococci. N. Engl. J. Med. 316, 927–931 (1987).
Gagnon, R. F., Richards, G. K. & Subang, R. Vancomycin therapy of experimental peritoneal catheter-associated infection (Staphylococcus epidermidis) in a mouse model. Perit. Dial. Int. 13 (Suppl. 2), 310–312 (1993).
Richards, G. K., Prentis, J. & Gagnon, R. F. Antibiotic activity against Staphylococcus epidermidis biofilms in dialysis fluids. Adv. Perit. Dial. 5, 133–137 (1989).
Raad, I. et al. Comparative activities of daptomycin, linezolid, and tigecycline against catheter-related methicillin-resistant Staphylococcus bacteremic isolates embedded in biofilm. Antimicrob. Agents Chemother. 51, 1656–1660 (2007).
Hanssen, A. M., Kjeldsen, G. & Sollid, J. U. Local variants of staphylococcal cassette chromosome mec in sporadic methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci: evidence of horizontal gene transfer? Antimicrob. Agents Chemother. 48, 285–296 (2004).
Chambers, H. F. The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 7, 178–182 (2001).
Diep, B. A. et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367, 731–739 (2006).
DeLeo, F. R. & Otto, M. An antidote for Staphylococcus aureus pneumonia? J. Exp. Med. 205, 271–274 (2008).
Otto, M. Targeted immunotherapy for staphylococcal infections: focus on anti-MSCRAMM antibodies. BioDrugs 22, 27–36 (2008).
Marraffini, L. A. & Sontheimer, E. J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845 (2008). This article describes the functional characterization of CRISPR sequences in S. epidermidis.
Rupp, M. E., Fey, P. D., Heilmann, C. & Gotz, F. Characterization of the importance of Staphylococcus epidermidis autolysin and polysaccharide intercellular adhesin in the pathogenesis of intravascular catheter-associated infection in a rat model. J. Infect. Dis. 183, 1038–1042 (2001).
Pintens, V. et al. The role of σB in persistence of Staphylococcus epidermidis foreign body infection. Microbiology 154, 2827–2836 (2008).
Vandecasteele, S. J., Peetermans, W. E., Merckx, R. & Van Eldere, J. Expression of biofilm-associated genes in Staphylococcus epidermidis during in vitro and in vivo foreign body infections. J. Infect. Dis. 188, 730–737 (2003).
Vuong, C., Kocianova, S., Yu, J., Kadurugamuwa, J. L. & Otto, M. Development of real-time in vivo imaging of device-related Staphylococcus epidermidis infection in mice and influence of animal immune status on susceptibility to infection. J. Infect. Dis. 198, 258–261 (2008).
Novick, R. P. & Geisinger, E. Quorum sensing in staphylococci. Annu. Rev. Genet. 42, 541–564 (2008).
Novick, R. P. et al. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12, 3967–3975 (1993).
Queck, S. Y. et al. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol. Cell 32, 150–158 (2008).
Mayville, P. et al. Structure–activity analysis of synthetic autoinducing thiolactone peptides from Staphylococcus aureus responsible for virulence. Proc. Natl Acad. Sci. USA 96, 1218–1223 (1999).
Otto, M., Sussmuth, R., Jung, G. & Gotz, F. Structure of the pheromone peptide of the Staphylococcus epidermidis agr system. FEBS Lett. 424, 89–94 (1998).
Otto, M. Staphylococcus aureus and Staphylococcus epidermidis peptide pheromones produced by the accessory gene regulator agr system. Peptides 22, 1603–1608 (2001).
Otto, M., Echner, H., Voelter, W. & Gotz, F. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69, 1957–1960 (2001).
Vuong, C., Gotz, F. & Otto, M. Construction and characterization of an agr deletion mutant of Staphylococcus epidermidis. Infect. Immun. 68, 1048–1053 (2000).
Simons, J. W. et al. Cloning, purification and characterisation of the lipase from Staphylococcus epidermidis — comparison of the substrate selectivity with those of other microbial lipases. Eur. J. Biochem. 253, 675–683 (1998).
Farrell, A. M., Foster, T. J. & Holland, K. T. Molecular analysis and expression of the lipase of Staphylococcus epidermidis. J. Gen. Microbiol. 139, 267–277 (1993).
Longshaw, C. M., Farrell, A. M., Wright, J. D. & Holland, K. T. Identification of a second lipase gene, gehD, in Staphylococcus epidermidis: comparison of sequence with those of other staphylococcal lipases. Microbiology 146, 1419–1427 (2000).
Lindsay, J. A., Riley, T. V. & Mee, B. J. Production of siderophore by coagulase-negative staphylococci and its relation to virulence. Eur. J. Clin. Microbiol. Infect. Dis. 13, 1063–1066 (1994).
Cotton, J. L., Tao, J. & Balibar, C. J. Identification and characterization of the Staphylococcus aureus gene cluster coding for staphyloferrin A. Biochemistry 48, 1025–1035 (2009).
Cockayne, A. et al. Molecular cloning of a 32-kilodalton lipoprotein component of a novel iron-regulated Staphylococcus epidermidis ABC transporter. Infect. Immun. 66, 3767–3774 (1998).
Chamberlain, N. R. & Brueggemann, S. A. Characterisation and expression of fatty acid modifying enzyme produced by Staphylococcus epidermidis. J. Med. Microbiol. 46, 693–697 (1997).
Park, P. W., Rosenbloom, J., Abrams, W. R., Rosenbloom, J. & Mecham, R. P. Molecular cloning and expression of the gene for elastin-binding protein (ebpS) in Staphylococcus aureus. J. Biol. Chem. 271, 15803–15809 (1996).
Acknowledgements
This work was supported by the intramural research programme of the National Institute of Allergy and Infectious Diseases.
Author information
Authors and Affiliations
Related links
Related links
DATABASES
Entrez Genome Project
FURTHER INFORMATION
Glossary
- Biofilm
-
A multicellular agglomeration of microorganisms that forms on a surface. Biofilms have a characteristic three-dimensional structure and physiology.
- Quorum sensing
-
A method of cell density-dependent gene regulation in bacteria. Quorum sensing systems in Gram-positive bacteria commonly contain peptide-based secreted signals and a membrane-located sensor. The staphylococcal quorum sensing system is termed agr and controls a series of genes involved in metabolism and virulence.
- Antimicrobial peptide
-
A peptide such as a defensin or cathelicidin, which have antimicrobial activity. Antimicrobial peptides are secreted by the host, for example, by epithelial cells or into neutrophil phagosomes.
- Innate host defence
-
A part of the immune system that provides the first line of defence, a fast response to invading microorganisms, based on recognition of pathogen-associated molecular patterns. The innate immune system consists mainly of phagocytes, platelets and secreted antimicrobial peptides.
- Neutrophil
-
The most abundant leukocyte in human blood. Neutrophils are the main cells that eliminate invading microorganisms by uptake and subsequent killing through reactive oxygen species and antimicrobial proteins and peptides.
- Acquired host defence
-
A part of the immune system that depends on antigen-dependent clonal expansion of T and B cells after antigen presentation by professional antigen-presenting cells. The acquired response provides long-term humoral (antibody-based) and cell-mediated immunity, but is delayed.
- Sortase
-
An enzyme that covalently links secreted bacterial surface proteins to peptidoglycan. Most of these proteins are substrates of sortase A and are characterized by an LPXTG amino acid motif at the carboxyl terminus.
- Teichoic acid
-
An anionic cell envelope glycopolymer produced by Gram-positive bacteria, composed of many identical sugar–phosphate-repeating units. Teichoic acids can be linked to peptidoglycan (wall teichoic acids) or to the cytoplasmic membrane through a lipid anchor (lipoteichoic acids).
- Pseudopeptide
-
A peptide that is formed by peptide bonds through carboxyl groups other than the α-carboxyl group.
- Pathogen-associated molecular pattern
-
A surface structure on pathogens that is recognized by the innate immune system as non-self and triggers activation of innate host defence, usually by binding to Toll-like receptors.
- Two-component system
-
A bacterial sensory system composed of a membrane-located sensor (histidine kinase) and a cytoplasmic DNA-binding regulatory protein (response regulator). The autophosphorylation-dependent activation of two-component systems is triggered by an extracellular signal.
- Enterotoxin
-
A protein toxin released by a microorganism into the intestine of its host.
- Methicillin
-
A penicillin derivative that is resistant to penicillinase (an enzyme widespread in staphylococci that provides resistance to penicillin).
- Mobile genetic element
-
DNA such as a plasmid or transposon that can be exchanged between bacteria by horizontal gene transfer. Mobile genetic elements often carry virulence or antibiotic resistance genes.
Rights and permissions
About this article
Cite this article
Otto, M. Staphylococcus epidermidis — the 'accidental' pathogen. Nat Rev Microbiol 7, 555–567 (2009). https://doi.org/10.1038/nrmicro2182
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2182