Key Points
-
Hepatitis C virus (HCV) infection is a major cause of chronic hepatitis, liver cirrhosis and hepatocellular carcinoma worldwide.
-
HCV is classified in the Hepacivirus genus within the Flaviviridae family. These enveloped positive-strand RNA viruses express their structural and non-structural proteins by the translation of a single long open reading frame.
-
HCV cell entry is a complex multistep process involving numerous cellular factors, including scavenger receptor class B type I, CD81 and claudin-1.
-
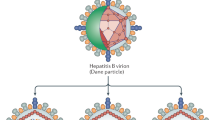
HCV structural proteins include the core protein and the envelope glycoproteins E1 and E2. The non-structural proteins include the p7 ion channel, the NS2–3 protease, the NS3 serine protease and RNA helicase, the NS4A polypeptide, the NS4B and NS5A proteins, and the NS5B RNA-dependent RNA polymerase.
-
HCV RNA replication takes place in a membrane-associated replication complex.
-
The recent development of complete cell-culture systems now allows the systematic dissection of the entire viral lifecycle.
Abstract
Exciting progress has recently been made in understanding the replication of hepatitis C virus, a major cause of chronic hepatitis, liver cirrhosis and hepatocellular carcinoma worldwide. The development of complete cell-culture systems should now enable the systematic dissection of the entire viral lifecycle, providing insights into the hitherto difficult-to-study early and late steps. These efforts have already translated into the identification of novel antiviral targets and the development of new therapeutic strategies, some of which are currently undergoing clinical evaluation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Williams, R. Global challenges in liver disease. Hepatology 44, 521–526 (2006).
Choo, Q.-L. et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244, 359–362 (1989).
Grakoui, A., McCourt, D. W., Wychowski, C., Feinstone, S. M. & Rice, C. M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J. Virol. 67, 2832–2843 (1993).
Bartenschlager, R., Ahlborn-Laake, L., Yasargil, K., Mous, J. & Jacobsen, H. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J. Virol. 68, 5045–5055 (1994).
Kolykhalov, A. A. et al. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277, 570–574 (1997).
Lohmann, V. et al. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285, 110–113 (1999).
Blight, K. J., Kolykhalov, A. A. & Rice, C. M. Efficient initiation of HCV RNA replication in cell culture. Science 290, 1972–1974 (2000).
Bartosch, B., Dubuisson, J. & Cosset, F. L. Infectious hepatitis C virus pseudo-particles containing functional E1–E2 envelope protein complexes. J. Exp. Med. 197, 633–642 (2003).
Hsu, M. et al. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl Acad. Sci. USA 100, 7271–7276 (2003).
Lindenbach, B. D. et al. Complete replication of hepatitis C virus in cell culture. Science 309, 623–626 (2005).
Wakita, T. et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nature Med. 11, 791–796 (2005).
Zhong, J. et al. Robust hepatitis C virus infection in vitro. Proc. Natl Acad. Sci. USA 102, 9294–9299 (2005).
Lindenbach, B. D. et al. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc. Natl Acad. Sci. USA 103, 3805–3809 (2006).
Pietschmann, T. et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl Acad. Sci. USA 103, 7408–7413 (2006).
Penin, F., Dubuisson, J., Rey, F. A., Moradpour, D. & Pawlotsky, J. M. Structural biology of hepatitis C virus. Hepatology 39, 5–19 (2004).
Lindenbach, B. D. & Rice, C. M. Unravelling hepatitis C virus replication from genome to function. Nature 436, 933–938 (2005).
Appel, N., Schaller, T., Penin, F. & Bartenschlager, R. From structure to function: new insights into hepatitis C virus RNA replication. J. Biol. Chem. 281, 9833–9836 (2006).
Moradpour, D. & Rice, C. M. in Hepatology. A textbook of liver disease (eds Boyer, T. D., Wright, T. L. & Manns, M. P.) 125–147 (Elsevier Science, 2006).
Thiel, H. J. et al. in Virus taxonomy. VIIIth Report of the International Committee on Taxonomy of Viruses (eds Fauquet, C. M., Mayo, M. A., Maniloff, J., Desselberger, U. & Ball, L. A.) 979–996 (Academic Press, 2005).
Neumann, A. U. et al. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-α therapy. Science 282, 103–107 (1998).
Simmonds, P. et al. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42, 962–973 (2005).
Kuiken, C. et al. Hepatitis C databases, principles and utility to researchers. Hepatology 43, 1157–1165 (2006).
Moradpour, D. et al. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J. Virol. 78, 7400–7409 (2004).
Lohmann, V., Körner, F., Dobierzewska, A. & Bartenschlager, R. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75, 1437–1449 (2001).
Blight, K. J., McKeating, J. A. & Rice, C. M. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76, 13001–13014 (2002).
Lohmann, V., Hoffmann, S., Herian, U., Penin, F. & Bartenschlager, R. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77, 3007–3019 (2003).
Bukh, J. et al. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc. Natl Acad. Sci. USA 99, 14416–14421 (2002).
Kato, T. et al. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125, 1808–1817 (2003).
Yi, M., Villanueva, R. A., Thomas, D. L., Wakita, T. & Lemon, S. M. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl Acad. Sci. USA 103, 2310–2315 (2006).
Kuhn, R. J. et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108, 717–725 (2002).
André, P., Perlemuter, G., Budkowska, A., Bréchot, C. & Lotteau, V. Hepatitis C virus particles and lipoprotein metabolism. Semin. Liver Dis. 25, 93–104 (2005).
Pileri, P. et al. Binding of hepatitis C virus to CD81. Science 282, 938–941 (1998).
Agnello, V., Abel, G., Elfahal, M., Knight, G. B. & Zhang, Q. X. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl Acad. Sci. USA 96, 12766–12771 (1999).
Scarselli, E. et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21, 5017–5025 (2002).
Evans, M. J. et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446, 801–805 (2007).
Bartosch, B. & Cosset, F. L. Cell entry of hepatitis C virus. Virology 348, 1–12 (2006).
Cocquerel, L., Voisset, C. & Dubuisson, J. Hepatitis C virus entry: potential receptors and their biological functions. J. Gen. Virol. 87, 1075–1084 (2006).
Coyne, C. B. & Bergelson, J. M. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell 124, 119–131 (2006).
Blanchard, E. et al. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 80, 6964–6972 (2006).
Koutsoudakis, G. et al. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 80, 5308–5320 (2006).
Tscherne, D. M. et al. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80, 1734–1741 (2006).
Modis, Y., Ogata, S., Clements, D. & Harrison, S. C. Structure of the dengue virus envelope protein after membrane fusion. Nature 427, 313–319 (2004).
Gibbons, D. L. et al. Conformational change and protein–protein interactions of the fusion protein of Semliki Forest virus. Nature 427, 320–325 (2004).
Friebe, P., Lohmann, V., Krieger, N. & Bartenschlager, R. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75, 12047–12057 (2001).
Jopling, C. L., Yi, M., Lancaster, A. M., Lemon, S. M. & Sarnow, P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309, 1577–1581 (2005).
Kolykhalov, A. A., Feinstone, S. M. & Rice, C. M. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70, 3363–3371 (1996).
Tanaka, T., Kato, N., Cho, M.-J., Sugiyama, K. & Shimotohno, K. Structure of the 3′ terminus of the hepatitis C virus genome. J. Virol. 70, 3307–3312 (1996).
Friebe, P. & Bartenschlager, R. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J. Virol. 76, 5326–5338 (2002).
Yi, M. & Lemon, S. M. 3′ nontranslated RNA signals required for replication of hepatitis C virus RNA. J. Virol. 77, 3557–3568 (2003).
Yanagi, M., St Claire, M., Emerson, S. U., Purcell, R. H. & Bukh, J. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl Acad. Sci. USA 96, 2291–2295 (1999).
Kolykhalov, A. A., Mihalik, K., Feinstone, S. M. & Rice, C. M. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J. Virol. 74, 2046–2051 (2000).
You, S., Stump, D. D., Branch, A. D. & Rice, C. M. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J. Virol. 78, 1352–1366 (2004).
Friebe, P., Boudet, J., Simorre, J.-P. & Bartenschlager, R. A kissing loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J. Virol. 79, 380–392 (2005).
Otto, G. A. & Puglisi, J. D. The pathway of HCV IRES-mediated translation initiation. Cell 119, 369–380 (2004).
Kieft, J. S., Zhou, K., Grech, A., Jubin, R. & Doudna, J. A. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nature Struct. Biol. 9, 370–374 (2002).
Lukavsky, P. J., Kim, I., Otto, G. A. & Puglisi, J. D. Structure of HCV IRES domain II determined by NMR. Nature Struct. Biol. 10, 1033–1038 (2003).
Spahn, C. M. et al. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science 291, 1959–1962 (2001).
Siridechadilok, B., Fraser, C. S., Hall, R. J., Doudna, J. A. & Nogales, E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310, 1513–1515 (2005).
McLauchlan, J., Lemberg, M. K., Hope, G. & Martoglio, B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21, 3980–3988 (2002).
Boulant, S. et al. Structural determinants that target the hepatitis C virus core protein to lipid droplets. J. Biol. Chem. 281, 22236–22247 (2006).
Asselah, T., Rubbia-Brandt, L., Marcellin, P. & Negro, F. Steatosis in chronic hepatitis C: why does it really matter? Gut 55, 123–130 (2006).
Dubuisson, J., Penin, F. & Moradpour, D. Interaction of hepatitis C virus proteins with host cell membranes and lipids. Trends Cell Biol. 12, 517–523 (2002).
Carrère-Kremer, S. et al. Subcellular localization and topology of the p7 polypeptide of hepatitis C virus. J. Virol. 76, 3720–3730 (2002).
Sakai, A. et al. The p7 polypeptide of hepatitis C virus is critical for infectivity and contains functionally important genotype-specific sequences. Proc. Natl Acad. Sci. USA 100, 11646–11651 (2003).
Griffin, S. D. et al. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett. 535, 34–38 (2003).
Pavlovic, D. et al. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl Acad. Sci. USA 100, 6104–6108 (2003).
Grakoui, A., McCourt, D. W., Wychowski, C., Feinstone, S. M. & Rice, C. M. A second hepatitis C virus-encoded proteinase. Proc. Natl Acad. Sci. USA 90, 10583–10587 (1993).
Hijikata, M. et al. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J. Virol. 67, 4665–4675 (1993).
Pallaoro, M. et al. Characterization of the hepatitis C virus NS2/3 processing reaction by using a purified precursor protein. J. Virol. 75, 9939–9946 (2001).
Thibeault, D., Maurice, R., Pilote, L., Lamarre, D. & Pause, A. In vitro characterization of a purified NS2/3 protease variant of hepatitis C virus. J. Biol. Chem. 276, 46678–46684 (2001).
Lorenz, I. C., Marcotrigiano, J., Dentzer, T. G. & Rice, C. M. Structure of the catalytic domain of the hepatitis C virus NS2–3 protease. Nature 442, 831–835 (2006).
Kalinina, O., Norder, H., Mukomolov, S. & Magnius, L. O. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 76, 4034–4043 (2002).
Noppornpanth, S. et al. Identification of a naturally occurring recombinant genotype 2/6 hepatitis C virus. J. Virol. 80, 7569–7577 (2006).
Yao, N., Reichert, P., Taremi, S. S., Prosise, W. W. & Weber, P. C. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure Fold. Des. 7, 1353–1363 (1999).
Wölk, B. et al. Subcellular localization, stability and trans-cleavage competence of the hepatitis C virus NS3–NS4A complex expressed in tetracycline-regulated cell lines. J. Virol. 74, 2293–2304 (2000).
Lamarre, D. et al. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426, 186–189 (2003).
Li, K. et al. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl Acad. Sci. USA 102, 2992–2997 (2005).
Meylan, E. et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437, 1167–1172 (2005).
Seth, R. B., Sun, L., Ea, C. K. & Chen, Z. J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122, 669–682 (2005).
Kawai, T. et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nature Immunol. 6, 981–988 (2005).
Xu, L. G. et al. VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol. Cell 19, 727–740 (2005).
Johnson, C. L. & Gale, M., Jr. CARD games between virus and host get a new player. Trends Immunol. 27, 1–4 (2006).
Serebrov, V. & Pyle, A. M. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature 430, 476–480 (2004).
Levin, M. K., Gurjar, M. & Patel, S. S. A Brownian motor mechanism of translocation and strand separation by hepatitis C virus helicase. Nature Struct. Mol. Biol. 12, 429–435 (2005).
Dumont, S. et al. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature 439, 105–108 (2006).
Kwong, A. D., Rao, B. G. & Jeang, K. T. Viral and cellular RNA helicases as antiviral targets. Nature Rev. Drug Discov. 4, 845–853 (2005).
Frick, D. N., Rypma, R. S., Lam, A. M. & Gu, B. The nonstructural protein 3 protease/helicase requires an intact protease domain to unwind duplex RNA efficiently. J. Biol. Chem. 279, 1269–1280 (2004).
Egger, D. et al. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76, 5974–5984 (2002).
Yu, G. Y., Lee, K. J., Gao, L. & Lai, M. M. Palmitoylation and polymerization of hepatitis C virus NS4B protein. J. Virol. 80, 6013–6023 (2006).
Quintavalle, M., Sambucini, S., Di Pietro, C., De Francesco, R. & Neddermann, P. The α-isoform of protein kinase CKI is responsible for hepatitis C virus NS5A hyperphosphorylation. J. Virol. 80, 11305–11312 (2006).
Reed, K. E., Gorbalenya, A. E. & Rice, C. M. The NS5A/NS5 proteins of viruses from three genera of the family flaviviridae are phosphorylated by associated serine/threonine kinases. J. Virol. 72, 6199–6206 (1998).
Evans, M. J., Rice, C. M. & Goff, S. P. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc. Natl Acad. Sci. USA 101, 13038–13043 (2004).
Neddermann, P. et al. Reduction of hepatitis C virus NS5A hyperphosphorylation by selective inhibition of cellular kinases activates viral RNA replication in cell culture. J. Virol. 78, 13306–13314 (2004).
Appel, N., Pietschmann, T. & Bartenschlager, R. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J. Virol. 79, 3187–3194 (2005).
Gao, L., Aizaki, H., He, J. W. & Lai, M. M. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J. Virol. 78, 3480–3488 (2004).
Penin, F. et al. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 279, 40835–40843 (2004).
Tellinghuisen, T. L., Marcotrigiano, J., Gorbalenya, A. E. & Rice, C. M. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J. Biol. Chem. 279, 48576–48587 (2004).
Tellinghuisen, T. L., Marcotrigiano, J. & Rice, C. M. Structure of the zinc-binding domain of an essential replicase component of hepatitis C virus reveals a novel fold. Nature 435, 375–379 (2005).
Huang, L. et al. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J. Biol. Chem. 280, 36417–36428 (2005).
Miyanari, Y. et al. Hepatitis C virus non-structural proteins in the probable membranous compartment function in viral RNA replication. J. Biol. Chem. 278, 50301–50308 (2003).
Quinkert, D., Bartenschlager, R. & Lohmann, V. Quantitative analysis of the hepatitis C virus replication complex. J. Virol. 79, 13594–13605 (2005).
Ago, H. et al. Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Struct. Fold. Des. 7, 1417–1426 (1999).
Lesburg, C. A. et al. Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nature Struct. Biol. 6, 937–943 (1999).
Bressanelli, S., Tomei, L., Rey, F. A. & De Francesco, R. Structural analysis of the hepatitis C virus RNA polymerase in complex with ribonucleotides. J. Virol. 76, 3482–3492 (2002).
Butcher, S. J., Grimes, J. M., Makeyev, E. V., Bamford, D. H. & Stuart, D. I. A mechanism for initiating RNA-dependent RNA polymerization. Nature 410, 235–240 (2001).
Lyle, J. M., Bullitt, E., Bienz, K. & Kirkegaard, K. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science 296, 2218–2222 (2002).
Wang, Q. M. et al. Oligomerization and cooperative RNA synthesis activity of hepatitis C virus RNA-dependent RNA polymerase. J. Virol. 76, 3865–3872 (2002).
Moradpour, D. et al. Membrane association of the RNA-dependent RNA polymerase is essential for hepatitis C virus RNA replication. J. Virol. 78, 13278–13284 (2004).
Branch, A. D., Stump, D. D., Gutierrez, J. A., Eng, F. & Walewski, J. L. The hepatitis C virus alternate reading frame (ARF) and its family of novel products: the alternate reading frame protein/F-protein, the double-frameshift protein, and others. Semin. Liver Dis. 25, 105–117 (2005).
McMullan, L. K. et al. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc. Natl Acad. Sci. USA 104, 2879–2884 (2007).
Salonen, A., Ahola, T. & Kääriäinen, L. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285, 139–173 (2004).
Mackenzie, J. Wrapping things up about virus RNA replication. Traffic 6, 967–977 (2005).
Schwartz, M. et al. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol. Cell 9, 505–514 (2002).
Gosert, R. et al. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J. Virol. 77, 5487–5492 (2003).
Ye, J. et al. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc. Natl Acad. Sci. USA 100, 15865–15870 (2003).
Kapadia, S. B. & Chisari, F. V. Hepatitis C virus RNA replication is regulated by host geranylgeranylation and fatty acids. Proc. Natl Acad. Sci. USA 102, 2561–2566 (2005).
Wang, C. et al. Identification of FBL2 as a geranylgeranylated cellular protein required for hepatitis C virus RNA replication. Mol. Cell 18, 425–434 (2005).
Sakamoto, H. et al. Host sphingolipid biosynthesis as a target for hepatitis C virus therapy. Nature Chem. Biol. 1, 333–337 (2005).
Watashi, K. et al. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell 19, 111–122 (2005).
Paeshuyse, J. et al. The non-immunosuppressive cyclosporin DEBIO-025 is a potent inhibitor of hepatitis C virus replication in vitro. Hepatology 43, 761–770 (2006).
Okamoto, T. et al. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 25, 5015–5025 (2006).
De Francesco, R. & Migliaccio, G. Challenges and successes in developing new therapies for hepatitis C. Nature 436, 953–960 (2005).
Trozzi, C. et al. In vitro selection and characterization of hepatitis C virus serine protease variants resistant to an active-site peptide inhibitor. J. Virol. 77, 3669–3679 (2003).
Yi, M. et al. Mutations conferring resistance to SCH6, a novel hepatitis C virus NS3/4A protease inhibitor. Reduced RNA replication fitness and partial rescue by second-site mutations. J. Biol. Chem. 281, 8205–8215 (2006).
Acknowledgements
Research in the authors' laboratories is supported by the Swiss National Science Foundation, the Swiss Cancer League/Oncosuisse, the Leenaards Foundation, the European Commission, the French Centre National de la Recherche Scientifique, the Agence Nationale de Recherche sur le SIDA et les Hépatites Virales, the US Public Health Service, the Greenberg Medical Research Institute, the Starr Foundation and the Ellison Medical Foundation. We dedicate this Review to the memory of Glovanni Migliaccio, a wonderful scientist and friend, who made many contributions to the HCV field.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
C.M.R. declares an equity interest in Apath, LLC, which holds commercial rights to the Huh-7.5 cells and other HCV-related technology.
Related links
Related links
DATABASES
Entrez Genome
Protein Data Bank
FURTHER INFORMATION
The Japanese Hepatitis Virus Database
Glossary
- Viral half-life
-
The time taken for half of the viruses to be cleared.
- Permissive cell
-
A cell that can be infected by, or supports the replication of, a virus.
- Fulminant hepatitis C
-
Fulminant hepatic failure refers to the rapid development of severe acute liver injury, with impaired synthetic function and encephalopathy, in a person who previously had a normal liver or had well-compensated liver disease.
- Pseudoknot
-
A pseudoknot is an RNA secondary structure containing two stem-loop structures in which the first stem's loop forms part of the second stem.
Rights and permissions
About this article
Cite this article
Moradpour, D., Penin, F. & Rice, C. Replication of hepatitis C virus. Nat Rev Microbiol 5, 453–463 (2007). https://doi.org/10.1038/nrmicro1645
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1645
This article is cited by
-
Targeting ER stress/PKA/GSK-3β/β-catenin pathway as a potential novel strategy for hepatitis C virus-infected patients
Cell Communication and Signaling (2023)
-
PPP2R5D promotes hepatitis C virus infection by binding to viral NS5B and enhancing viral RNA replication
Virology Journal (2022)
-
Sofosbuvir-based direct-acting antivirals and changes in cholesterol and low density lipoprotein-cholesterol
Scientific Reports (2022)
-
Partial nonstructural 3 region analysis of hepatitis C virus genotype 3a
Molecular Biology Reports (2022)
-
Effect of sofosbuvir-based DAAs on changes in lower-density lipoprotein in HCV patients: a systematic review and meta-analysis
BMC Infectious Diseases (2021)