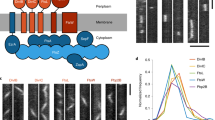
Key Points
-
All cells must divide to proliferate, and most bacteria divide by splitting themselves into two during cytokinesis. Many bacteria divide by splitting into approximately equal halves in a process called binary fission. Cytokinesis in bacteria is achieved by the divisome, a dedicated protein machine that is located at the site of cell division. Recent advances in ultrastructural imaging, biochemistry and genetics of Escherichia coli and other model bacterial species have helped to refine models of divisome function and regulation.
-
FtsZ, the bacterial homologue of tubulin, is the principal driver of bacterial cytokinesis. In vitro, FtsZ assembles into single protofilaments in the presence of GTP. In vivo, these protofilaments loosely assemble to encircle the cell at the site of division — called the Z ring — and are positioned there by species-specific spatial positioning proteins.
-
As FtsZ is a soluble protein, FtsZ protofilaments must be tethered to the inner surface of the cytoplasmic membrane by additional proteins, including FtsA and ZipA in E. coli. This complex of FtsZ and membrane tethers is called the proto-ring and has highly dynamic behaviour.
-
Although they do not form microtubules, FtsZ protofilaments self-associate to form bundles, either through interactions with other FtsZ subunits or with several FtsZ-binding proteins that enhance bundling, including ZipA and Zap proteins. These lateral interactions between FtsZ protofilaments may be important for the ability of FtsZ to divide a cell.
-
FtsA, a bacterial homologue of actin, is a key connector between the Z ring and other proteins of the divisome, all of which span the membrane and some of which bind to the peptidoglycan layer. Once the divisome is completely assembled, it coordinates the inward constriction of the Z ring and cytoplasmic membrane with the synthesis of the cell division septum, which is composed of peptidoglycan. FtsA is a key player in this coordination, which probably involves feedback signalling between the peptidoglycan-binding divisome proteins and the Z ring. Biochemical characterization of FtsA remains a major challenge.
-
In addition to signalling in the divisome during the process of cytokinesis, the divisome is regulated by mechanical, metabolic and stress inputs. FtsZ is a major target for these regulators, but other divisome proteins are also targets. Understanding how divisome proteins are inhibited or stimulated will be valuable in the future design of divisome-specific antimicrobial compounds.
Abstract
Bacteria must divide to increase in number and colonize their niche. Binary fission is the most widespread means of bacterial cell division, but even this relatively simple mechanism has many variations on a theme. In most bacteria, the tubulin homologue FtsZ assembles into a ring structure, termed the Z ring, at the site of cytokinesis and recruits additional proteins to form a large protein machine — the divisome — that spans the membrane. In this Review, we discuss current insights into the regulation of the assembly of the Z ring and how the divisome drives membrane invagination and septal cell wall growth while flexibly responding to various cellular inputs.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
25 April 2016
Nature Reviews Microbiology 14, 305–319 (2016). In the sixth paragraph of the section 'FtsA and the dynamics of proto-ring assembly', the sentence “Finally, an FtsZ mutant with decreased self-bundling in vitro (FtsZ-E93R) has reduced function in cell division50, 51.” should have read “However, an FtsZ mutant with increased self-bundling in vitro (FtsZ-E93R) has reduced function in cell division50,51.” The authors apologize to the readers for any misunderstanding caused.
References
Egan, A. J. & Vollmer, W. The physiology of bacterial cell division. Ann. NY Acad. Sci. 1277, 8–28 (2013).
Brown, P. J. et al. Polar growth in the alphaproteobacterial order Rhizobiales. Proc. Natl Acad. Sci. USA 109, 1697–1701 (2011).
Margolin, W. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24, 531–548 (2000).
Leisch, N. et al. Growth in width and FtsZ ring longitudinal positioning in a gammaproteobacterial symbiont. Curr. Biol. 22, R831–R832 (2012).
Monahan, L. G., Liew, A. T., Bottomley, A. L. & Harry, E. J. Division site positioning in bacteria: one size does not fit all. Front. Microbiol. 5, 19 (2014).
Rowlett, V. W. & Margolin, W. The Min system and other nucleoid-independent regulators of Z ring positioning. Front. Microbiol. 6, 478 (2015).
Willemse, J., Borst, J. W., de Waal, E., Bisseling, T. & van Wezel, G. P. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev. 25, 89–99 (2011).This study provides the first example of positive spatial regulation of Z ring positioning.
Treuner-Lange, A. et al. PomZ, a ParA-like protein, regulates Z-ring formation and cell division in Myxococcus xanthus. Mol. Microbiol. 87, 235–253 (2013).
Holeckova, N. et al. LocZ is a new cell division protein involved in proper septum placement in Streptococcus Pneumoniae. mBio 6, e01700-14 (2014).
Fleurie, A. et al. MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature 516, 259–262 (2014).
McCormick, J. R., Su, E. P., Driks, A. & Losick, R. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol. Microbiol. 14, 243–254 (1994).
Rico, A. I., Krupka, M. & Vicente, M. In the beginning. Escherichia coli assembled the proto-ring: an initial phase of division. J. Biol. Chem. 288, 20830–20836 (2013).
Pichoff, S. & Lutkenhaus, J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 55, 1722–1734 (2005).
Hale, C. A. & de Boer, P. A. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88, 175–185 (1997).
Pichoff, S. & Lutkenhaus, J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21, 685–693 (2002).
Duman, R. et al. Structural and genetic analyses reveal the protein SepF as a new membrane anchor for the Z ring. Proc. Natl Acad. Sci. USA 110, E4601–E4610 (2013).
Krol, E. et al. Bacillus subtilis SepF binds to the C-terminus of FtsZ. PLoS ONE 7, e43293 (2013).
Ishikawa, S., Kawai, Y., Hiramatsu, K., Kuwano, M. & Ogasawara, N. A new FtsZ-interacting protein, YlmF, complements the activity of FtsA during progression of cell division in Bacillus subtilis. Mol. Microbiol. 60, 1364–1380 (2006).
Gupta, S. et al. The essential protein SepF of mycobacteria interacts with FtsZ and MurG to regulate cell growth and division. Microbiology 161, 1627–1638 (2015).
Gola, S., Munder, T., Casonato, S., Manganelli, R. & Vicente, M. The essential role of SepF in mycobacterial division. Mol. Microbiol. 97, 560–576 (2015).
Levin, P. A., Kurtser, I. G. & Grossman, A. D. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl Acad. Sci. USA 96, 9642–9647 (1999).
Steele, V. R., Bottomley, A. L., Garcia-Lara, J., Kasturiarachchi, J. & Foster, S. J. Multiple essential roles for EzrA in cell division of Staphylococcus aureus. Mol. Microbiol. 80, 542–555 (2011).
Haeusser, D. P. et al. EzrA prevents aberrant cell division by modulating assembly of the cytoskeletal protein FtsZ. Mol. Microbiol. 52, 801–814 (2004).
Cleverley, R. M. et al. Structure and function of a spectrin-like regulator of bacterial cytokinesis. Nat. Commun. 5, 5421 (2014).
Machnicka, B. et al. Spectrins: a structural platform for stabilization and activation of membrane channels, receptors and transporters. Biochim. Biophys. Acta 1838, 620–634 (2014).
Haeusser, D. P., Garza, A. C., Buscher, A. Z. & Levin, P. A. The division inhibitor EzrA contains a seven-residue patch required for maintaining the dynamic nature of the medial FtsZ ring. J. Bacteriol. 189, 9001–9010 (2007).
Land, A. D., Luo, Q. & Levin, P. A. Functional domain analysis of the cell division inhibitor EzrA. PLoS ONE 9, e102616 (2014).
Son, S. H. & Lee, H. H. The N-terminal domain of EzrA binds to the C terminus of FtsZ to inhibit Staphylococcus aureus FtsZ polymerization. Biochem. Biophys. Res. Commun. 433, 108–114 (2013).
van den Ent, F. & Löwe, J. Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J. 19, 5300–5307 (2000).
Sanchez, M., Valencia, A., Ferrandiz, M. J., Sandler, C. & Vicente, M. Correlation between the structure and biochemical activities of FtsA, an essential cell division protein of the actin family. EMBO J. 13, 4919–4925 (1994).
Feucht, A., Lucet, I., Yudkin, M. D. & Errington, J. Cytological and biochemical characterization of the FtsA cell division protein of Bacillus subtilis. Mol. Microbiol. 40, 115–125 (2001).
Paradis-Bleau, C., Sanschagrin, F. & Levesque, R. C. Peptide inhibitors of the essential cell division protein FtsA. Protein Eng. Des. Sel. 18, 85–91 (2005).
Lara, B. et al. Cell division in cocci: localization and properties of the Streptococcus pneumoniae FtsA protein. Mol. Microbiol. 55, 699–711 (2005).The first study to demonstrate that FtsA has ATP-dependent polymerization activity.
Szwedziak, P., Wang, Q., Freund, S. M. & Löwe, J. FtsA forms actin-like protofilaments. EMBO J. 31, 2249–2260 (2012).This study structurally defines the FtsA–FtsA and FtsA–FtsZ interfaces.
Pichoff, S., Shen, B., Sullivan, B. & Lutkenhaus, J. FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA's self-interaction competes with its ability to recruit downstream division proteins. Mol. Microbiol. 83, 151–167 (2012).This study shows that many mutations that interfere with FtsA oligomerization render ZipA non-essential.
Shiomi, D. & Margolin, W. Dimerization or oligomerization of the actin-like FtsA protein enhances the integrity of the cytokinetic Z ring. Mol. Microbiol. 66, 1396–1415 (2007).
Fujita, J. et al. Crystal structure of FtsA from Staphylococcus aureus. FEBS Lett. 588, 1879–1885 (2014).
Geissler, B., Elraheb, D. & Margolin, W. A gain of function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc. Natl Acad. Sci. USA 100, 4197–4202 (2003).
Geissler, B. & Margolin, W. Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Mol. Microbiol. 58, 596–612 (2005).
Beuria, T. K. et al. Adenine nucleotide-dependent regulation of assembly of bacterial tubulin-like FtsZ by a hypermorph of bacterial actin-like FtsA. J. Biol. Chem. 284, 14079–14086 (2009).The first evidence that FtsA can affect the assembly state of FtsZ.
Loose, M. & Mitchison, T. J. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat. Cell Biol. 16, 38–46 (2014).This study shows that FtsA can regulate FtsZ assembly dynamics on a physiological membrane surface.
Modi, K. & Misra, H. S. Dr-FtsA, an actin homologue in Deinococcus radiodurans differentially affects Dr-FtsZ and Ec-FtsZ functions in vitro. PLoS ONE 9, e115918 (2014).
Du, S., Park, K.-T. & Lutkenhaus, J. Oligomerization of FtsZ converts the FtsZ tail motif (CCTP) into a multivalent ligand with high avidity for partners ZipA and SlmA. Mol. Microbiol. 95, 173–188 (2015).
Herricks, J. R., Nguyen, D. & Margolin, W. A thermosensitive defect in the ATP binding pocket of FtsA can be suppressed by allosteric changes in the dimer interface. Mol. Microbiol. 94, 713–727 (2014).
Shen, B. & Lutkenhaus, J. The conserved C-terminal tail of FtsZ is required for the septal localization and division inhibitory activity of MinCC/MinD. Mol. Microbiol. 72, 410–424 (2009).
Rowlett, V. W. & Margolin, W. Asymmetric constriction of dividing Escherichia coli cells induced by expression of a fusion between two Min proteins. J. Bacteriol. 196, 2089–2100 (2014).
Szwedziak, P., Wang, Q., Bharat, T. A. M., Tsim, M. & Löwe, J. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife 3, e04601 (2014).
Hernandez-Rocamora, V. M. et al. Dynamic interaction of the Escherichia coli cell division ZipA and FtsZ proteins evidenced in nanodiscs. J. Biol. Chem. 287, 30097–30104 (2012).
Skoog, K. & Daley, D. O. The Escherichia coli cell division protein ZipA forms homodimers prior to association with FtsZ. Biochemistry 51, 1407–1415 (2012).
Jaiswal, R. et al. E93R substitution of Escherichia coli FtsZ induces bundling of protofilaments, reduces GTPase activity, and impairs bacterial cytokinesis. J. Biol. Chem. 285, 31796–31805 (2010).
Haeusser, D. P., Rowlett, V. W. & Margolin, W. A mutation in Escherichia coli ftsZ bypasses the requirement for the essential division gene zipA and confers resistance to FtsZ assembly inhibitors by stabilizing protofilament bundling. Mol. Microbiol. 97, 988–1005 (2015).
Dewar, S. J., Begg, K. J. & Donachie, W. D. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J. Bacteriol. 174, 6314–6316 (1992).
Mukherjee, A. & Lutkenhaus, J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 17, 462–469 (1998).
Scheffers, D. J., de Wit, J. G., den Blaauwen, T. & Driessen, A. J. GTP hydrolysis of cell division protein FtsZ: evidence that the active site is formed by the association of monomers. Biochemistry 41, 521–529 (2002).
Huang, K. H., Durand-Heredia, J. & Janakiraman, A. FtsZ ring stability: of bundles, tubules, crosslinks, and curves. J. Bacteriol. 195, 1859–1868 (2013).
Meier, E. L. & Goley, E. D. Form and function of the bacterial cytokinetic ring. Curr. Opin. Cell Biol. 26, 147 (2014).
Rowlett, V. W. & Margolin, W. The bacterial divisome: ready for its close-up. Phil. Trans. R. Soc. Lond. B Biol. Sci. 370, 20150028 (2015).
Li, Z., Trimble, M. J., Brun, Y. V. & Jensen, G. J. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 26, 4694–4708 (2007).This study provides a glimpse of the Z ring at high resolution by electron tomography.
Holden, S. J. et al. High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc. Natl Acad. Sci. USA 111, 4566–4571 (2014).
Jacq, M. et al. Remodeling of the Z-ring nanostructure during the Streptococcus pneumoniae cell cycle revealed by photoactivated localization microscopy. mBio 6, e01108-15 (2015).
Strauss, M. P. et al. 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Biol. 10, e1001389 (2012).This paper provides the direct visual evidence that the Z ring may be a non-uniform structure.
Rowlett, V. W. & Margolin, W. 3D-SIM super-resolution of FtsZ and its membrane tethers in Escherichia coli cells. Biophys. J. 107, L17–L20 (2014).
Tsui, H. C. T. et al. Pbp2x localizes separately from Pbp2b and other peptidoglycan synthesis proteins during later stages of cell division of Streptococcus pneumoniae D39. Mol. Microbiol. 94, 21–40 (2014).
Piro, O., Carmon, G., Feingold, M. & Fishov, I. Three-dimensional structure of the Z-ring as a random network of FtsZ filaments. Env. Microbiol. 15, 3252–3258 (2013).
Si, F., Busiek, K., Margolin, W. & Sun, S. X. Organization of FtsZ filaments in the bacterial division ring measured from polarized fluorescence microscopy. Biophys. J. 105, 1976–1986 (2013).
Judd, E. M. et al. Distinct constrictive processes, separated in time and space, divide Caulobacter inner and outer membranes. J. Bacteriol. 187, 6874–6882 (2005).
Gundogdu, M. E. et al. Large ring polymers align FtsZ polymers for normal septum formation. EMBO J. 30, 617–626 (2011).
Hale, C. A., Rhee, A. C. & de Boer, P. A. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol. 182, 5153–5166 (2000).
Roach, E. J., Kimber, M. S. & Khursigara, C. M. Crystal structure and site-directed mutational analysis reveals key residues involved in Escherichia coli ZapA function. J. Biol. Chem. 289, 23276–23286 (2014).
Durand-Heredia, J. M., Yu, H. H., De Carlo, S., Lesser, C. F. & Janakiraman, A. Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli. J. Bacteriol. 193, 1405–1413 (2011).
Hale, C. A. et al. Identification of Escherichia coli ZapC (YcbW) as a component of the division apparatus that binds and bundles FtsZ polymers. J. Bacteriol. 193, 1393–1404 (2011).
Durand-Heredia, J., Rivkin, E., Fan, G., Morales, J. & Janakiraman, A. Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J. Bacteriol. 194, 3189–3198 (2012).
Buss, J. et al. In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol. Microbiol. 89, 1099–1120 (2013).
Gardner, K. A., Moore, D. A. & Erickson, H. P. The C-terminal linker of Escherichia coli FtsZ functions as an intrinsically disordered peptide. Mol. Microbiol. 89, 264–275 (2013).
Buske, P. J. & Levin, P. A. A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Mol. Microbiol. 89, 249–263 (2013).
Sundararajan, K. et al. The bacterial tubulin FtsZ requires its intrinsically disordered linker to direct robust cell wall construction. Nat. Commun. 6, 7281 (2015).
Buske, P. J. & Levin, P. A. Extreme C-terminus of bacterial cytoskeletal protein FtsZ plays fundamental role in assembly independent of modulatory proteins. J. Biol. Chem. 287, 10945–10957 (2012).
Fu, G. et al. in vivo structure of the E. coli FtsZ-ring revealed by photoactivated localization microscopy (PALM). PLoS ONE 5, e12682 (2010).
Aarsman, M. E. et al. Maturation of the Escherichia coli divisome occurs in two steps. Mol. Microbiol. 55, 1631–1645 (2005).
Gamba, P., Veening, J. W., Saunders, N. J., Hamoen, L. W. & Daniel, R. A. Two-step assembly dynamics of the Bacillus subtilis divisome. J. Bacteriol. 191, 4186–4194 (2009).
Mohammadi, T. et al. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 30, 1425–1432 (2011).
Sham, L. T. et al. Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345, 220–222 (2014).
Van den Berg van Saparoea, H. B. et al. Fine-mapping the contact sites of the Escherichia coli cell division proteins FtsB and FtsL on the FtsQ protein. J. Biol. Chem. 288, 24340–24350 (2013).
Khadria, A. S. & Senes, A. The transmembrane domains of the bacterial cell division proteins FtsB and FtsL form a stable high-order oligomer. Biochemistry 52, 7542–7550 (2013).
Glas, M. et al. The soluble periplasmic domains of E. coli cell division proteins FtsQ/FtsB/FtsL form a trimeric complex with sub-micromolar affinity. J. Biol. Chem. 290, 21498–21509 (2015).
Bottomley, A. L. et al. Staphylococcus aureus DivIB is a peptidoglycan-binding protein that is required for a morphological checkpoint in cell division. Mol. Microbiol. 94, 1041–1064 (2014).
Grenga, L., Rizzo, A., Paolozzi, L. & Ghelardini, P. Essential and non-essential interactions in interactome networks: the Escherichia coli division proteins FtsQ–FtsN interaction. Env. Microbiol. 15, 3210–3217 (2013).
Alexeeva, S. et al. Direct interactions of early and late assembling division proteins in Escherichia coli cells resolved by FRET. Mol. Microbiol. 77, 384–398 (2010).
Busiek, K. K., Eraso, J. M., Wang, Y. & Margolin, W. The early divisome protein FtsA interacts directly through its 1c subdomain with the cytoplasmic domain of the late divisome protein FtsN. J. Bacteriol. 194, 1989–2000 (2012).
Rico, A. I., Garcia-Ovalle, M., Mingorance, J. & Vicente, M. Role of two essential domains of Escherichia coli FtsA in localization and progression of the division ring. Mol. Microbiol. 53, 1359–1371 (2004).
Yahashiri, A., Jorgenson, M. A. & Weiss, D. S. Bacterial SPOR domains are recruited to septal peptidoglycan by binding to glycan strands that lack stem peptides. Proc. Natl Acad. Sci. USA 112, 11347–11352 (2015).
Gerding, M. A. et al. Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J. Bacteriol. 191, 7383–7401 (2009).
Busiek, K. K. & Margolin, W. A role for FtsA in SPOR-independent localization of the essential Escherichia coli cell division protein FtsN. Mol. Microbiol. 92, 1212–1226 (2014).
Bernard, C. S., Sadasivam, M., Shiomi, D. & Margolin, W. An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli. Mol. Microbiol. 64, 1289–1305 (2007).
Liu, B., Persons, L., Lee, L. & de Boer, P. Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol. Microbiol. 95, 945–970 (2015).Together with references 89, 93 and 97, this study provides additional evidence for several signalling pathways in the divisome to activate cytokinesis.
Pichoff, S., Du, S. & Lutkenhaus, J. The bypass of ZipA by overexpression of FtsN requires a previously unknown conserved FtsN motif essential for FtsA–FtsN interaction supporting a model in which FtsA monomers recruit late cell division proteins to the Z ring. Mol. Microbiol. 95, 971–987 (2015).
Tsang, M. J. & Bernhardt, T. G. A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol. Microbiol. 95, 925–944 (2015).
Potluri, L. P., Kannan, S. & Young, K. D. ZipA is required for FtsZ-dependent preseptal peptidoglycan synthesis prior to invagination during cell division. J. Bacteriol. 194, 5334–5342 (2012).
Claessen, D. et al. Control of the cell elongation–division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol. Microbiol. 68, 1029–1046 (2008).
Land, A. D. et al. Requirement of essential Pbp2x and GpsB for septal ring closure in Streptococcus pneumoniae D39. Mol. Microbiol. 90, 939–955 (2013).
Fleurie, A. et al. Interplay of the serine/threonine-kinase StkP and the paralogs DivIVA and GpsB in pneumococcal cell elongation and division. PLoS Genet. 10, e1004275 (2014).
Pompeo, F., Foulquier, E., Serrano, B., Grangeasse, C. & Galinier, A. Phosphorylation of the cell division protein GpsB regulates PrkC kinase activity through a negative feedback loop in Bacillus subtilis. Mol. Microbiol. 97, 139–150 (2015).
Gerding, M. A., Ogata, Y., Pecora, N. D., Niki, H. & de Boer, P. A. The trans-envelope Tol–Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli. Mol. Microbiol. 63, 1008–1025 (2007).The first mechanism for coordinating inner membrane and outer membrane constriction during cytokinesis in a Gram-negative bacterium.
Gray, A. N. et al. Coordination of peptidoglycan synthesis and outer membrane constriction during Escherichia coli cell division. eLife 4, e07118 (2015).
Corbin, B. D., Wang, Y., Beuria, T. K. & Margolin, W. Interaction between cell division proteins FtsE and FtsZ. J. Bacteriol. 189, 3026–3035 (2007).
Yang, D. C. et al. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc. Natl Acad. Sci. USA 108, E1052–E1060 (2011).
Jorgenson, M. A., Chen, Y., Yahashiri, A., Popham, D. L. & Weiss, D. S. The bacterial septal ring protein RlpA is a lytic transglycosylase that contributes to rod shape and daughter cell separation in Pseudomonas aeruginosa. Mol. Microbiol. 93, 113–128 (2014).
Osawa, M., Anderson, D. E. & Erickson, H. P. Reconstitution of contractile FtsZ rings in liposomes. Science 320, 792–794 (2008).The first study to demonstrate that Z rings can be reconstituted in vitro.
Osawa, M. & Erickson, H. P. Liposome division by a simple bacterial division machinery. Proc. Natl Acad. Sci. USA 110, 11000–11004 (2013).This study shows that purified FtsZ and FtsA* are sufficient to constrict liposomes.
Dow, C. E., van den Berg, H. A., Roper, D. I. & Rodger, A. Biological insights from a simulation model of the critical FtsZ accumulation required for prokaryotic cell division. Biochemistry 54, 3803–3813 (2015).
Li, Y. et al. FtsZ protofilaments use a hinge-opening mechanism for constrictive force generation. Science 341, 392–395 (2013).This paper proposes a mechanism for force generation by FtsZ filaments attached to membranes.
Söderström, B. et al. Disassembly of the divisome in Escherichia coli: evidence that FtsZ dissociates before compartmentalization. Mol. Microbiol. 92, 1–9 (2014).
Krupka, M. et al. Role of the FtsA C-terminus as a switch for polymerization and membrane association. mBio 5, e02221 (2014).This study further defines the role of ATP binding for FtsA activity.
Cabre, E. J. et al. Bacterial division proteins FtsZ and ZipA induce vesicle shrinkage and cell membrane invagination. J. Biol. Chem. 288, 26625–26634 (2013).
Weiss, D. S. Last but not least: new insights into how FtsN triggers constriction during Escherichia coli cell division. Mol. Microbiol. 903–909 (2015).
Tsang, M. J. & Bernhardt, T. G. Guiding divisome assembly and controlling its activity. Curr. Opin. Microbiol. 24, 60–65 (2015).
Weart, R. B. et al. A metabolic sensor governing cell size in bacteria. Cell 130, 335–347 (2007).This study is the first to demonstrate a mechanism for metabolic regulation of bacterial cell division.
Chien, A. C., Zareh, S. K., Wang, Y. M. & Levin, P. A. Changes in the oligomerization potential of the division inhibitor UgtP co-ordinate Bacillus subtilis cell size with nutrient availability. Mol. Microbiol. 86, 594–610 (2012).
Hill, N. S., Buske, P. J., Shi, Y. & Levin, P. A. A moonlighting enzyme links Escherichia coli cell size with central metabolism. PLoS Genet. 9, e1003663 (2013).
Surdova, K. et al. The conserved DNA-binding protein WhiA is involved in cell division in Bacillus subtilis. J. Bacteriol. 195, 5450–5460 (2013).
Monahan, L. G., Hajduk, I. V., Blaber, S. P., Charles, I. G. & Harry, E. J. Coordinating bacterial cell division with nutrient availability: a role for glycolysis. mBio 5, e00935-14 (2014).
Radhakrishnan, S. K., Pritchard, S. & Viollier, P. H. Coupling prokaryotic cell fate and division control with a bifunctional and oscillating oxidoreductase homolog. Dev. Cell 18, 90–101 (2010).
Beaufay, F. et al. A NAD-dependent glutamate dehydrogenase coordinates metabolism with cell division in Caulobacter crescentus. EMBO J. 34, 1786–1800 (2015).
Takada, H. et al. An essential enzyme for phospholipid synthesis associates with the Bacillus subtilis divisome. Mol. Microbiol. 91, 242–255 (2014).
Weart, R. B., Nakano, S., Lane, B. E., Zuber, P. & Levin, P. A. The ClpX chaperone modulates assembly of the tubulin-like protein FtsZ. Mol. Microbiol. 57, 238–249 (2005).
Camberg, J. L., Hoskins, J. R. & Wickner, S. The interplay of ClpXP with the cell division machinery in Escherichia coli. J. Bacteriol. 193, 1911–1918 (2011).
Williams, B., Bhat, N., Chien, P. & Shapiro, L. ClpXP and ClpAP proteolytic activity on divisome substrates is differentially regulated following the Caulobacter asymmetric cell division. Mol. Microbiol. 93, 853–866 (2014).
Chen, Y., Milam, S. L. & Erickson, H. P. SulA inhibits assembly of FtsZ by a simple sequestration mechanism. Biochemistry 51, 3100–3109 (2012).
Modell, J. W., Hopkins, A. C. & Laub, M. T. DNA damage checkpoint in Caulobacter crescentus inhibits cell division through a direct interaction with FtsW. Genes Dev. 25, 1328–1343 (2011).
Modell, J. W., Kambara, T. K., Perchuk, B. S. & Laub, M. T. DNA damage-induced, SOS-independent checkpoint regulates cell division in Caulobacter crescentus. PLoS Biol. 12, e1001977 (2014).
Mo, A. H. & Burkholder, W. F. YneA, an SOS-induced inhibitor of cell division in Bacillus subtilis, is regulated posttranslationally and requires the transmembrane region for activity. J. Bacteriol. 192, 3159–3173 (2010).
Buchholz, M., Nahrstedt, H., Pillukat, M. H., Deppe, V. & Meinhardt, F. yneA mRNA instability is involved in temporary inhibition of cell division during the SOS response of Bacillus megaterium. Microbiology 159, 1564–1574 (2013).
Marteyn, B. S. et al. ZapE is a novel cell division protein interacting with FtsZ and modulating the Z-ring dynamics. mBio 5, e00022-14 (2014).
Koprowski, P. et al. Cytoplasmic domain of MscS interacts with cell division protein FtsZ: A possible non-channel function of the mechanosensitive channel in Escherichia coli. PLoS ONE 10, e0127029 (2015).
Wilson, M. E., Jensen, G. S. & Haswell, E. S. Two mechanosensitive channel homologs influence division ring placement in Arabidopsis chloroplasts. Plant Cell 23, 2939–2949 (2011).
Mercier, R., Kawai, Y. & Errington, J. General principles for the formation and proliferation of a wall-free (L-form) state in bacteria. eLife 3, e04629 (2014).
Lock, R. L. & Harry, E. J. Cell-division inhibitors: new insights for future antibiotics. Nat. Rev. Drug Discov. 7, 324–338 (2008).
Ojima, I., Kumar, K., Awasthi, D. & Vineberg, J. G. Drug discovery targeting cell division proteins, microtubules and FtsZ. Bioorg. Med. Chem. 22, 5060–5077 (2014).
Sass, P. & Brotz-Oesterhelt, H. Bacterial cell division as a target for new antibiotics. Curr. Opin. Microbiol. 16, 522–530 (2013).
Artola, M. et al. Effective GTP-replacing FtsZ inhibitors and antibacterial mechanism of action. ACS Chem. Biol. 10, 834–843 (2014).
Haeusser, D. P. et al. The Kil peptide of bacteriophage λ blocks Escherichia coli cytokinesis via ZipA-dependent inhibition of FtsZ assembly. PLoS Genet. 10, e1004217 (2014).
Kiro, R. et al. Gene product 0.4 increases bacteriophage T7 competitiveness by inhibiting host cell division. Proc. Natl Acad. Sci. USA 110, 19549–19554 (2013).
Hernandez-Rocamora, V. M., Alfonso, C., Margolin, W., Zorrilla, S. & Rivas, G. Evidence that bacteriophage λ Kil peptide inhibits bacterial cell division by disrupting FtsZ protofilaments and sequestering protein subunits. J. Biol. Chem. 290, 20325–20335 (2015).
Ballesteros-Plaza, D., Holguera, I., Scheffers, D. J., Salas, M. & Munoz-Espin, D. Phage ϕ29 protein p1 promotes replication by associating with the FtsZ ring of the divisome in Bacillus subtilis. Proc. Natl Acad. Sci. USA 110, 12313–12318 (2013).
Bisson-Filho, A. W. et al. FtsZ filament capping by MciZ, a developmental regulator of bacterial division. Proc. Natl Acad. Sci. USA 112, E2130–E2138 (2015).
Pende, N. et al. Size-independent symmetric division in extraordinarily long cells. Nat. Commun. 5, 4803 (2014).
Donovan, C. & Bramkamp, M. Cell division in Corynebacterineae. Front. Microbiol. 5, 132 (2014).
Kieser, K. J. & Rubin, E. J. How sisters grow apart: mycobacterial growth and division. Nat. Rev. Microbiol. 12, 550–562 (2014).
Ramos-Leon, F., Mariscal, V., Frias, J. E., Flores, E. & Herrero, A. Divisome-dependent subcellular localization of cell-cell joining protein SepJ in the filamentous cyanobacterium Anabaena. Mol. Microbiol. 96, 566–580 (2015).
Pinho, M. G., Kjos, M. & Veening, J.-W. How to get (a)round: mechanisms controlling growth and division of coccoid bacteria. Nat. Rev. Microbiol. 11, 601–614 (2013).
Angert, E. R. Alternatives to binary fission in bacteria. Nat. Rev. Microbiol. 3, 214–224 (2005).
Tuson, H. H. & Biteen, J. S. Unveiling the inner workings of live bacteria using super-resolution microscopy. Anal. Chem. 87, 42–63 (2015).
Asano, S., Engel, B. D. & Baumeister, W. In situ cryo-electron tomography: A post-reductionist approach to structural biology. J. Mol. Biol. 428, 332–343 (2015).
Busiek, K. K. & Margolin, W. Bacterial actin and tubulin homologs in cell growth and division. Curr. Biol. 25, 245–254 (2015).
Grainge, I. FtsK–a bacterial cell division checkpoint? Mol. Microbiol. 78, 1055–1057 (2010).
Buss, J. et al. A multi-layered protein network stabilizes the Escherichia coli FtsZ-ring and modulates constriction dynamics. PLoS Genet. 11, e1005128 (2015).
Acknowledgements
The authors gratefully acknowledge support from the US National Institute of General Medical Sciences (R01-GM61074 to W.M.) and the US National Institute of Research Resources (S10RR029552) for the use of the 3D-SIM microscope.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Glossary
- Cytokinesis
-
The splitting of the contents of a cell to make two cells.
- Nucleoids
-
The nucleus-like organized structures of bacterial chromosomes.
- Mycelium
-
A filamentous, branched network of multinucleate cells growing on a surface.
- Lipid II flippase
-
A membrane protein that transfers lipid-linked peptidoglycan precursors from the inner leaflet of the cytoplasmic membrane to the outer leaflet of the cytoplasmic membrane, so that they can be incorporated into the cell wall.
- Sidewall
-
The peptidoglycan layer in many rod-shaped bacteria that is active in elongating the cell and comprises most of the straight wall of the cell with the exception of cell poles and the division septum.
- Thermosensitive ftsZ mutant
-
A point mutation in the ftsZ gene that permits normal growth and division at 30 °C but stops division at 42 °C despite continued growth, resulting in filamentous, multinucleate cells (hence the term fts for filamentous temperature sensitive mutants).
- Central metabolism
-
Metabolic pathways, such as the tricarboxylic acid (TCA) cycle, that provide precursor metabolites for all other pathways that are required for growth.
- ClpXP
-
A protein chaperone–protease machine that targets certain proteins for unfolding and degradation.
- SOS response
-
Induced by DNA damage, the SOS response is a stress response that results in the expression of many genes that are important for the protection of chromosomal DNA.
Rights and permissions
About this article
Cite this article
Haeusser, D., Margolin, W. Splitsville: structural and functional insights into the dynamic bacterial Z ring. Nat Rev Microbiol 14, 305–319 (2016). https://doi.org/10.1038/nrmicro.2016.26
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro.2016.26
This article is cited by
-
Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody
Nature Communications (2023)
-
Bacterial cell-size changes resulting from altering the relative expression of Min proteins
Nature Communications (2023)
-
Application of nanotags and nanobodies for live cell single-molecule imaging of the Z-ring in Escherichia coli
Current Genetics (2023)
-
Assembly dynamics of FtsZ and DamX during infection-related filamentation and division in uropathogenic E. coli
Nature Communications (2022)
-
Function and regulation of the divisome for mitochondrial fission
Nature (2021)