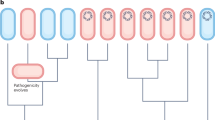
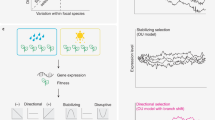
Abstract
Orthologues and paralogues are types of homologous genes that are related by speciation or duplication, respectively. Orthologous genes are generally assumed to retain equivalent functions in different organisms and to share other key properties. Several recent comparative genomic studies have focused on testing these expectations. Here we discuss the complexity of the evolution of gene–phenotype relationships and assess the validity of the key implications of orthology and paralogy relationships as general statistical trends and guiding principles.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Fitch, W. M. Distinguishing homologous from analogous proteins. Systemat. Zool. 19, 99–106 (1970).
Fitch, W. M. Homology a personal view on some of the problems. Trends Genet. 16, 227–231 (2000).
Koonin, E. V. Walter Fitch and the orthology paradigm. Brief. Bioinform. 12, 377–378 (2011).
Baldauf, S. L. Phylogeny for the faint of heart: a tutorial. Trends Genet. 19, 345–351 (2003).
Sonnhammer, E. L. & Koonin, E. V. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 18, 619–620 (2002).
Koonin, E. V. Orthologs, paralogs and evolutionary genomics. Annu. Rev. Genet. 39, 309–338 (2005).
Dolinski, K. & Botstein, D. Orthology and functional conservation in eukaryotes. Annu. Rev. Genet. 41, 465–507 (2007).
Studer, R. A. & Robinson-Rechavi, M. How confident can we be that orthologs are similar, but paralogs differ? Trends Genet. 25, 210–216 (2009).
Nehrt, N. L., Clark, W. T., Radivojac, P. & Hahn, M. W. Testing the ortholog conjecture with comparative functional genomic data from mammals. PLoS Comput. Biol. 7, e1002073 (2011).
Kuzniar, A., van Ham, R. C., Pongor, S. & Leunissen, J. A. The quest for orthologs: finding the corresponding gene across genomes. Trends Genet. 24, 539–551 (2008).
Gabaldón, T. Large-scale assignment of orthology: back to phylogenetics? Genome Biol. 9, 235 (2008).
Altenhoff, A. M. & Dessimoz, C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput. Biol. 5, e1000262 (2009).
Trachana, K. et al. Orthology prediction methods: a quality assessment using curated protein families. Bioessays 33, 769–780 (2011).
Kristensen, D. M., Wolf, Y. I., Mushegian, A. R. & Koonin, E. V. Computational methods for Gene Orthology inference. Brief. Bioinform. 12, 379–391 (2011).
Gabaldón, T. et al. Joining forces in the quest for orthologs. Genome Biol. 10, 403 (2009).
Dessimoz, C., Gabaldón, T., Roos, D. S., Sonnhammer, E. L. & Herrero, J. Toward community standards in the quest for orthologs. Bioinformatics 28, 900–904 (2012).
Descorps-Declere, S., Lemoine, F., Sculo, Q., Lespinet, O. & Labedan, B. The multiple facets of homology and their use in comparative genomics to study the evolution of genes, genomes, and species. Biochimie 90, 595–608 (2008).
Mahmood, K., Webb, G. I., Song, J., Whisstock, J. C. & Konagurthu, A. S. Efficient large-scale protein sequence comparison and gene matching to identify orthologs and co-orthologs. Nucleic Acids Res. 40, e44 (2012).
Roux, J. & Robinson-Rechavi, M. An ontology to clarify homology-related concepts. Trends Genet. 26, 99–102 (2010).
Tatusov, R. L., Koonin, E. V. & Lipman, D. J. A genomic perspective on protein families. Science 278, 631–637 (1997).
Remm, M., Storm, C. E. & Sonnhammer, E. L. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 314, 1041–1052 (2001).
Koonin, E. V., Aravind, L. & Kondrashov, A. S. The impact of comparative genomics on our understanding of evolution. Cell 101, 573–576 (2000).
Sjolander, K., Datta, R. S., Shen, Y. & Shoffner, G. M. Ortholog identification in the presence of domain architecture rearrangement. Brief. Bioinform. 12, 413–422 (2011).
Forslund, K. & Sonnhammer, E. L. Evolution of protein domain architectures. Methods Mol. Biol. 856, 187–216 (2012).
Hartmann, B. & Valcarcel, J. Decrypting the genome's alternative messages. Curr. Opin. Cell Biol. 21, 377–386 (2009).
Irimia, M. & Blencowe, B. J. Alternative splicing: decoding an expansive regulatory layer. Curr. Opin. Cell Biol. 24, 323–332 (2012).
Basu, M. K., Poliakov, E. & Rogozin, I. B. Domain mobility in proteins: functional and evolutionary implications. Brief. Bioinform. 10, 205–216 (2009).
Ouzounis, C. Orthology: another terminology muddle. Trends Genet. 15, 445 (1999).
Theissen, G. Birth, life and death of developmental control genes: new challenges for the homology concept. Theory Biosci. 124, 199–212 (2005).
Bandyopadhyay, S., Sharan, R. & Ideker, T. Systematic identification of functional orthologs based on protein network comparison. Genome Res. 16, 428–435 (2006).
Singh, R., Xu, J. & Berger, B. Global alignment of multiple protein interaction networks with application to functional orthology detection. Proc. Natl Acad. Sci. USA 105, 12763–12768 (2008).
Huynen, M. A. & Bork, P. Measuring genome evolution. Proc. Natl Acad. Sci. USA 95, 5849–5856 (1998).
Bromham, L. & Penny, D. The modern molecular clock. Nature Rev. Genet. 4, 216–224 (2003).
Kumar, S. Molecular clocks: four decades of evolution. Nature Rev. Genet. 6, 654–662 (2005).
Koski, L. B. & Golding, G. B. The closest BLAST hit is often not the nearest neighbor. J. Mol. Evol. 52, 540–542 (2001).
Hulsen, T., Huynen, M. A., de Vlieg, J. & Groenen, P. M. Benchmarking ortholog identification methods using functional genomics data. Genome Biol. 7, R31 (2006).
Wolf, Y. I. & Koonin, E. V. A tight link between orthologs and bidirectional best hits in bacterial and archaeal genomes. Genome Biol. Evol. 1286–1294 (2012).
Koonin, E. V. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nature Rev. Microbiol. 1, 127–136 (2003).
Snel, B., Huynen, M. A. & Dutilh, B. E. Genome trees and the nature of genome evolution. Annu. Rev. Microbiol. 59, 191–209 (2005).
Blomme, T. et al. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 7, R43 (2006).
Makarova, K. S., Wolf, Y. I., Mekhedov, S. L., Mirkin, B. G. & Koonin, E. V. Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acids Res. 33, 4626–4638 (2005).
Huerta-Cepas, J. & Gabaldón, T. Assigning duplication events to relative temporal scales in genome-wide studies. Bioinformatics 27, 38–45 (2011).
Forslund, K., Pekkari, I. & Sonnhammer, E. L. Domain architecture conservation in orthologs. BMC Bioinformatics 12, 326 (2011).
Koonin, E. V. et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 5, R7 (2004).
Peterson, M. E. et al. Evolutionary constraints on structural similarity in orthologs and paralogs. Protein Sci. 18, 1306–1315 (2009).
Mushegian, A. R. & Koonin, E. V. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl Acad. Sci. USA 93, 10268–10273 (1996).
Galperin, M. Y., Walker, D. R. & Koonin, E. V. Analogous enzymes: independent inventions in enzyme evolution. Genome Res. 8, 779–790 (1998).
Omelchenko, M. V., Galperin, M. Y., Wolf, Y. I. & Koonin, E. V. Non-homologous isofunctional enzymes: a systematic analysis of alternative solutions in enzyme evolution. Biol. Direct 5, 31 (2010).
Lynch, V. J. & Wagner, G. P. Resurrecting the role of transcription factor change in developmental evolution. Evolution 62, 2131–2154 (2008).
Casci, T. Functional genomics: Degrees of similarity. Nature Rev. Genet. 12, 522 (2011).
Thomas, P. D., Wood, V., Mungall, C. J., Lewis, S. E. & Blake, J. A. On the use of gene ontology annotations to assess functional similarity among orthologs and paralogs: a short report. PLoS Comput. Biol. 8, e1002386 (2012).
Altenhoff, A. M., Studer, R. A., Robinson-Rechavi, M. & Dessimoz, C. Resolving the ortholog conjecture: orthologs tend to be weakly, but significantly, more similar in function than paralogs. PLoS Comput. Biol. 8, e1002514 (2012).
Huerta-Cepas, J., Dopazo, J., Huynen, M. A. & Gabaldón, T. Evidence for short-time divergence and long-time conservation of tissue-specific expression after gene duplication. Brief. Bioinform. 12, 442–448 (2011).
Chen, X. & Zhang, J. The ortholog conjecture is untestable by the current gene ontology but is supported by RNA sequencing data. PLoS Comput. Biol. 8, e1002784 (2012).
Mohd-Padil, H., Mohd-Adnan, A. & Gabaldón, T. Phylogenetic analyses uncover a novel clade of transferring in non-mammalian vertebrates. Mol. Biol. Evol. 30, 894–905 (2013).
Huerta-Cepas, J. et al. PhylomeDB v3.0: an expanding repository of genome-wide collections of trees, alignments and phylogeny-based orthology and paralogy predictions. Nucleic Acids Res. 39, D556–D560 (2011).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Acknowledgements
T.G. is supported by funds from the European Research Council and the Spanish Ministry of Economy and Competitiveness. E.V.K. is supported by intramural funds of the US Department of Health and Human Services (to the US National Library of Medicine).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
PowerPoint slides
Glossary
- Alternative transcription
-
The expression of multiple transcripts with different structures from the same gene locus.
- Bidirectional best hit
-
(BBH). A pair of genes that show the greatest sequence similarity to each other in a complete, reciprocal comparison of the gene (protein) sequences from a pair of compared genomes.
- Co-orthologue
-
A gene in a species (or group of species) that is jointly orthologous to the same gene (or genes) in another species (or group of species).
- Domain accretion
-
In evolution, the addition of sequences encoding extra structural domains to protein-coding genes.
- Gene Ontology
-
(GO). A collaborative bioinformatic project aiming at providing an ontology of defined terms representing gene product properties.
- In-paralogues
-
Paralogous genes that originate from a lineage-specific duplication that postdates that reference ancestral species.
- Non-homologous and isofunctional
-
When referring to proteins, these are proteins that in different species carry out equivalent biological functions but are not homologous.
- Orthologues
-
Homologous genes related by speciation.
- Orthologous groups
-
Sets of genes that are inferred to have evolved from a single ancestral gene in the reference ancestral species.
- Out-paralogues
-
Paralogous genes that originate from a duplication that antedates that reference ancestral species.
- Paralogues
-
Homologous genes related by duplication.
- Xenologues
-
Homologous genes that originate from horizontal gene transfer.
Rights and permissions
About this article
Cite this article
Gabaldón, T., Koonin, E. Functional and evolutionary implications of gene orthology. Nat Rev Genet 14, 360–366 (2013). https://doi.org/10.1038/nrg3456
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrg3456
This article is cited by
-
From beer to breadboards: yeast as a force for biological innovation
Genome Biology (2024)
-
Characterization of the genetic variation and evolutionary divergence of the CLEC18 family
Journal of Biomedical Science (2024)
-
The G2-Like gene family in Populus trichocarpa: identification, evolution and expression profiles
BMC Genomic Data (2023)
-
In silico secretome analyses of the polyphagous root-knot nematode Meloidogyne javanica: a resource for studying M. javanica secreted proteins
BMC Genomics (2023)
-
Approaches to increase the validity of gene family identification using manual homology search tools
Genetica (2023)