Key Points
-
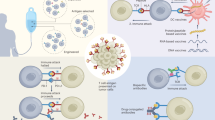
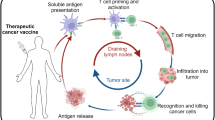
T-cell epitopes from tumour antigens have been included in many vaccination studies, and their potential to induce antitumour immune responses has become manifest. Nevertheless, the clinical outcome of such studies has to be improved.
-
The number of known epitopes is still limited; therefore, some tumours cannot be treated by immunotherapeutic approaches. For others, the efficacy is not yet optimal.
-
T-cell epitopes from tumour antigens can be defined by two principal strategies: one starts from an existing T-cell response and identifies the target of the response, whereas the other uses the sequence of a tumour antigen and employs epitope prediction to identify the relevant epitopes.
-
New tumour antigens can be discovered by analysing the specificity of existing T-cell responses, or by screening strategies such as the SEREX programme, comparative proteome analysis or gene-expression profiling.
-
To improve the clinical outcome of antitumour vaccinations, tumour-escape mechanisms have to be avoided by the use of efficient, multitarget vaccines.
-
With the growing number of T-cell epitopes, it will become feasible to design patient-specific, individual vaccines that address several different antigens from one tumour and several HLA specificities, including class-II-restricted epitopes.
Abstract
Ten years ago, the first melanoma patient was successfully treated by vaccination with a short peptide, which was, in fact, the first tumour-specific T-cell epitope ever defined — MAGE. Since then, a number of clinical vaccination studies have underlined the potential of tumour-specific T-cell epitopes. But, how can we identify more epitopes to improve their efficacy as an anticancer treatment?
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Renkvist, N., Castelli, C., Robbins, P. F. & Parmiani, G. A listing of human tumor antigens recognized by T cells. Cancer Immunol. Immunother. 50, 3–15 (2001).
Nestle, F. O. et al. Vaccination of melanoma patients with peptide- or tumor lysate-pulsed dendritic cells. Nature Med. 4, 328–332 (1998).
Rosenberg, S. A. et al. Immunologic and therapeutic evaluation of a synthetic peptide vaccine for the treatment of patients with metastatic melanoma. Nature Med. 4, 321–327 (1998).
Lee, K. H. et al. Increased vaccine-specific T cell frequency after peptide-based vaccination correlates with increased susceptibility to in vitro stimulation but does not lead to tumor regression. J. Immunol. 163, 6292–6300 (1999).
Panelli, M. C. et al. Phase I study in patients with metastatic melanoma of immunization with dendritic cells presenting epitopes derived from the melanoma-associated antigens MART1 and GP100. J. Immunother. 23, 487–498 (2000).
Bodey, B., Bodey, B. Jr, Siegel, S. E. & Kaiser, H. E. Failure of cancer vaccines: the significant limitations of this approach to immunotherapy. Anticancer Res. 20, 2665–2676 (2000).
Ohnmacht, G. A. et al. Short-term kinetics of tumor antigen expression in response to vaccination. J. Immunol. 167, 1809–1820 (2001).
Tait, B. D. HLA class I expression on human cancer cells. Implications for effective immunotherapy. Hum. Immunol. 61, 158–165 (2000).
Seliger, B. et al. Immune escape of melanoma: first evidence of structural alterations in two distinct components of the MHC class I antigen processing pathway. Cancer Res. 61, 8647–8650 (2001).
Rammensee, H.-G., Bachmann, J., Emmerich, N., Bachor, O. A. & Stevanovic, S. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50, 213–219 (1999).
Vonderheide, R. H. et al. Characterization of HLA-A3-restricted cytotoxic T lymphocytes reactive against the widely expressed tumor antigen telomerase. Clin. Cancer Res. 7, 3343–3348 (2001).An actual example of reverse immunology, using a ubiquitous tumour antigen, telomerase. Epitope identification was achieved after induction of peptide-specific T cells.
Schmitz, M. et al. Generation of survivin-specific CD8+ T effector cells by dendritic cells pulsed with protein or selected peptides. Cancer Res. 60, 4845–4849 (2000).
Mashino, K. et al. Expression of multiple cancer-testis antigen genes in gastrointestinal and breast carcinomas. Br. J. Cancer 85, 713–720 (2001).
Gillespie, A. M. et al. MAGE, BAGE and GAGE: tumour antigen expression in benign and malignant ovarian tissue. Br. J. Cancer 78, 816–821 (1998).
Russo, V. et al. Expression of the MAGE gene family in primary and metastatic human breast cancer: implications for tumor antigen-specific immunotherapy. Int. J. Cancer 64, 216–221 (1995).
van der Bruggen, P. et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 254, 1643–1647 (1991).
Traversari, C. et al. A nonapeptide encoded by human gene MAGE1 is recognized on HLA-A1 by cytolytic T lymphocytes directed against tumor antigen MZ2-E. J. Exp. Med. 176, 1453–1457 (1992).
Smith, E. S. et al. Lethality-based selection of recombinant genes in mammalian cells: application to identifying tumor antigens. Nature Med. 7, 967–972 (2001).A new generation of expression libraries that comprises enrichment steps.
Cox, A. L. et al. Identification of a peptide recognized by five melanoma-specific human cytotoxic T cell lines. Science 264, 716–719 (1994).
Falk, K., Rötzschke, O., Stevanovic, S., Jung, G. & Rammensee, H.-G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351, 290–296 (1991).
Rötzschke, O. et al. Exact prediction of a natural T cell epitope. Eur. J. Immunol. 21, 2891–2894 (1991).
Stevanovic, S. & Rammensee, H.-G. Identification of T-cell epitopes using allele-specific ligand motifs. Behring. Inst. Mitt. 95, 7–13 (1994).
Celis, E. et al. Induction of anti-tumor cytotoxic T lymphocytes in normal humans using primary cultures and synthetic peptide epitopes. Proc. Natl Acad. Sci. USA 91, 2105–2109 (1994).
Parker, K. C., Bednarek, M. A. & Coligan, J. E. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 152, 163–175 (1994).
Nussbaum, A. K. et al. PapRoC: a predicition algorithm for proteasomal cleavages available on the WWW. Immunogenetics 53, 87–94 (2001).
Kesmir, C. et al. 〈http://www.cbs.dtu.dk/services/NetChop/〉.
Schirle, M., Weinschenk, T. & Stevanovic, S. Combining computer algorithms with experimental approaches permits the rapid and accurate identification of T cell epitopes from defined antigens. J. Immunol. Methods 257, 1–16 (2001).
Kessler, J. H. et al. Efficient identification of novel HLA-A(*)0201-presented cytotoxic T lymphocyte epitopes in the widely expressed tumor antigen PRAME by proteasome-mediated digestion analysis. J. Exp. Med. 193, 73–88 (2001).Use of proteasomal digestion for the definition of HLA-presented peptides from tumour antigens.
Pascolo, S. et al. A MAGE-A1 HLA-A*0201 epitope identified by mass spectrometry. Cancer Res. 61, 4072–4077 (2001).Identification of a tumour-specific class I epitope by mass spectrometry without the use of pre-existing T cells.
Brossart, P. et al. Identification of HLA-A2 restricted T cell epitopes derived from the MUC1 tumor antigen for broadly applicable vaccine therapies. Blood 93, 4309–4317 (1999).
Schirle, M. et al. Identification of tumor-associated MHC class I ligands by a novel T cell-independent approach. Eur. J. Immunol. 30, 2216–2225 (2000).
Sahin, U. et al. Human neoplasms elicit multiple specific immune responses in the autologous host. Proc. Natl Acad. Sci. USA 92, 11810–11813 (1995).
Czerwenka, K. F. et al. Comparative analysis of two-dimensional protein patterns in malignant and normal human breast tissue. Cancer Detect. Prev. 25, 268–279 (2001).
Meehan, K. L., Holland, J. W. & Dawkins, H. J. Proteomic analysis of normal and malignant prostate tissue to identify novel proteins lost in cancer. Prostate 50, 54–63 (2002).
Emmert-Buck, M. R. et al. An approach to proteomic analysis of human tumors. Mol. Carcinog. 27, 158–165 (2000).
Mathiassen, S., Lauemöller, S. L., Ruhwald, M., Claesson, M. H. & Buus, S. Tumor-associated antigens identified by mRNA expression profiling induce protective anti-tumor immunity. Eur. J. Immunol. 31, 1239–1246 (2001).Candidate antigens from gene-expression profiling were used for epitope prediction, leading to protective T-cell epitopes in a mouse model.
Boer, J. M. et al. Identification and classification of differentially expressed genes in renal cell carcinoma by expression profiling on a global human 31,500-element cDNA array. Genome Res. 11, 1861–1870 (2001).
Klade C. S. et al. Identification of tumor antigens in renal cell carcinoma by serological proteome analysis. Proteomics 1, 890–898 (2001).
van Driel, W. J. et al. Vaccination with HPV16 peptides of patients with advanced cervical carcinoma: clinical evaluation of a Phase I–II trial. Eur. J. Cancer 35, 946–952 (1999).
Slingluff, C. L. Jr et al. Phase I trial of a melanoma vaccine with GP100(280–288) peptide and tetanus helper peptide in adjuvant: immunologic and clinical outcomes. Clin. Cancer Res. 7, 3012–3024 (2001).
Brossart, P. et al. Induction of cytotoxic T-lymphocyte responses in vivo after vaccinations with peptide-pulsed dendritic cells. Blood 96, 3102–3108 (2000).
Kobayashi, H., Song, Y., Hoon, D. S., Appella, E. & Celis, E. Tumor-reactive T helper lymphocytes recognize a promiscuous MAGE-A3 epitope presented by various major histocompatibility complex class II alleles. Cancer Res. 61, 4773–4778 (2001).A recent example of how to identify tumour-specific HLA class-II-presented epitopes.
Schultz, E. S. et al. A MAGE-A3 peptide presented by HLA-DP4 is recognized on tumor cells by CD4+ cytolytic T lymphocytes. Cancer Res. 60, 6272–6275 (2000).
Kobayashi, H., Lu, J. & Celis, E. Identification of helper T-cell epitopes that encompass or lie proximal to cytotoxic T-cell epitopes in the GP100 melanoma tumor antigen. Cancer Res. 61, 7577–7584 (2001).
Chaux, P. et al. MAGE1 peptide recognized on HLA-DR15 by CD4+ T cells. Eur. J. Immunol. 31, 1910–1916 (2001).
Casares, N. et al. Immunization with a tumor-associated CTL epitope plus a tumor-related or unrelated TH1 helper peptide elicits protective CTL immunity. Eur. J. Immunol. 31, 1780–1789 (2001).
Marks, M. S. et al. A lysosomal targeting signal in the cytoplasmic tail of the β-chain directs HLA-DM to MHC class II compartments. J. Cell Biol. 131, 351–369 (1995).
Sanderson, S., Frauwirth, K. & Shastri, N. Expression of endogenous peptide–major histocompatibility complex class II complexes derived from invariant chain-antigen fusion proteins. Proc. Natl Acad. Sci. USA 92, 7217–7221 (1995).
Malcherek, G. et al. MHC class II-associated invariant chain peptide replacement by T cell epitopes: engineered invariant chain as a vehicle for directed and enhanced MHC class II antigen processing and presentation. Eur. J. Immunol. 28, 1524–1533 (1998).
Rodriguez, F., Harkins, S., Redwine, J. M., de Pereda, J. M. & Whitton, J. L. CD4+ T cells induced by a DNA vaccine: immunological consequences of epitope-specific lysosomal targeting. J. Virol. 75, 10421–10430 (2001).
van der Bruggen, P. et al. A peptide encoded by human gene MAGE3 and presented by HLA-A2 induces cytolytic T lymphocytes that recognize tumor cells expressing MAGE3. Eur. J. Immunol. 24, 3038–3043 (1994).
Jäger, E. et al. Simultaneous humoral and cellular immune response against cancer-testis antigen NY-ESO-1: definition of human histocompatibility leukocyte antigen (HLA)-A2-binding peptide epitopes. J. Exp. Med. 187, 265–270 (1998).
Wölfel, T. et al. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science 269, 1281–1284 (1995).
Robbins, P. F. et al. A mutated β-catenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. J. Exp. Med. 183, 1185–1192 (1996).
Mandruzzato, S., Stroobant, V., Demotte, N. & van der Bruggen, P. A human CTL recognizes a caspase-8-derived peptide on autologous HLA-B*3503 molecules and two unrelated peptides on allogeneic HLA-B*3501 molecules. J. Immunol. 164, 4130–4134 (2000).
Ressing, M. E. et al. Human CTL epitopes encoded by human papillomavirus type 16 E6 and E7 identified through in vivo and in vitro immunogenicity studies of HLA-A*0201-binding peptides. J. Immunol. 154, 5934–5943 (1995).
Brichard, V. G. et al. A tyrosinase nonapeptide presented by HLA-B44 is recognized on a human melanoma by autologous cytolytic T lymphocytes. Eur. J. Immunol. 26, 224–230 (1996).
Skipper, J. C. et al. Shared epitopes for HLA-A3-restricted melanoma-reactive human CTL include a naturally processed epitope from PMEL17/GP100. J. Immunol. 157, 5027–5033 (1996).
Salgaller, M. L. et al. Dendritic cell-based immunotherapy of prostate cancer. Crit. Rev. Immunol. 18, 109–119 (1998).
Fisk, B., Blevins, T. L., Wharton, J. T. & Ioannides, C. G. Identification of an immunodominant peptide of HER2/neu protooncogene recognized by ovarian tumor-specific cytotoxic T lymphocyte lines. J. Exp. Med. 181, 2109–2117 (1995).
Kawashima, I. et al. Identification of HLA-A3-restricted cytotoxic T lymphocyte epitopes from carcinoembryonic antigen and HER2/neu by primary in vitro immunization with peptide-pulsed dendritic cells. Cancer Res. 59, 431–435 (1999).
Scheibenbogen, C. et al. Phase 2 trial of vaccination with tyrosinase peptides and granulocyte-macrophage colony-stimulating factor in patients with metastatic melanoma. J. Immunother. 23, 275–281 (2000).
Hunger, R. E. et al. Successful induction of immune responses against mutant RAS in melanoma patients using intradermal injection of peptides and GM–CSF as adjuvant. Exp. Dermatol. 10, 161–167 (2001).
Mackensen, A. et al. Phase I study in melanoma patients of a vaccine with peptide-pulsed dendritic cells generated in vitro from CD34+ hematopoietic progenitor cells. Int. J. Cancer 86, 385–392 (2000).
Yu, J. S. et al. Vaccination of malignant glioma patients with peptide-pulsed dendritic cells elicits systemic cytotoxicity and intracranial T-cell infiltration. Cancer Res. 61, 842–847 (2001).
Gjertsen, M. K. et al. Intradermal RAS peptide vaccination with granulocyte-macrophage colony-stimulating factor as adjuvant: clinical and immunological responses in patients with pancreatic adenocarcinoma. Int. J. Cancer 92, 441–450 (2001).
Gajewski, T. F., Fallarino, F., Ashikari, A. & Sherman, M. Immunization of HLA-A2+ melanoma patients with MAGE3 or MelanA peptide-pulsed autologous peripheral blood mononuclear cells plus recombinant human interleukin 12. Clin. Cancer Res. 7, 895s–901s (2001).
Schott, M. et al. Immunotherapy for medullary thyroid carcinoma by dendritic cell vaccination. J. Clin. Endocrinol. Metab. 86, 4965–4969 (2001).
Jäger, E. et al. Induction of primary NY-ESO-1 immunity: CD8+ T lymphocyte and antibody responses in peptide-vaccinated patients with NY-ESO-1+ cancers. Proc. Natl Acad. Sci. USA 97, 12198–12203 (2000).
Rosenberg, S. A. et al. Impact of cytokine administration on the generation of antitumor reactivity in patients with metastatic melanoma receiving a peptide vaccine. J. Immunol. 163, 1690–1695 (1999).
Murphy, G. P. et al. Infusion of dendritic cells pulsed with HLA-A2-specific prostate-specific membrane antigen peptides: a phase II prostate cancer vaccine trial involving patients with hormone-refractory metastatic disease. Prostate 38, 73–78 (1999).
Sadanaga, N. et al. Dendritic cell vaccination with MAGE peptide is a novel therapeutic approach for gastrointestinal carcinomas. Clin. Cancer Res. 7, 2277–2284 (2001).
Author information
Authors and Affiliations
Related links
Related links
DATABASES
Cancer.gov
LocusLink
Medscape DrugInfo
FURTHER INFORMATION
Proteasomal cleavage prediction NetChop
Glossary
- HLAs
-
Human leukocyte antigens, which are molecules of the human major histocompatibility complex.
- CD8
-
A surface molecule that is expressed exclusively by cytotoxic T cells.
- CD4
-
A surface molecule that is expressed exclusively by T-helper cells.
- MHC CLASS II MOLECULES
-
Peptide receptors, similar to class I molecules in structure and function, but exclusively expressed by a small set of immune cells. They mainly present peptides from extracellular proteins to T-helper cells.
- CYTOTOXIC T LYMPHOCYTES
-
(CTLs: killer cells, cytotoxic T cells). These control all major histocompatibility complex class-I-expressing body cells for the presence of abnormal (viral, tumour-associated) peptides.
- MHC CLASS I MOLECULES
-
Highly polymorphic glycoproteins that are expressed by every nucleated body cell of vertebrates, and that are encoded by the gene cluster 'major histocompatibility complex' (MHC). The human MHC molecules are termed HLA (human leukocyte antigen) molecules. MHC class I molecules mainly present peptides from intracellular proteins to cytotoxic T cells.
- DENDRITIC CELLS
-
(DCs). These present T-cell epitopes very efficiently and are able to activate cytotoxic T lymphocytes.
- MELANOMA
-
Skin cancer, originating from transformed melanocytes.
- MAGE
-
Melanoma antigen; a family of proteins that are expressed only in testis or tumour cells.
- PROTEOMICS
-
Analysis of the entirety of proteins from a tissue sample, separated by two-dimensional gel electrophoresis. Each individual protein spot can be identified after tryptic digestion and mass spectrometrical analysis of the resulting peptides.
- MICROARRAY ANALYSIS
-
'DNA chips' contain many thousands of oligonucleotides that represent fragments of genes, immobilized on a solid surface. They are probed with cDNA that is prepared from mRNA of tissue samples and so give quantitative information about gene expression.
- MELANOSOMES
-
Subcellular organelles in melanocytes, which contain the skin's pigments.
- PERIPHERAL-BLOOD MONONUCLEAR CELLS
-
(PBMCs). Includes B cells, T cells and monocytes. These can be obtained from whole blood after ficoll density-gradient centrifugation.
Rights and permissions
About this article
Cite this article
Stevanovic, S. Identification of tumour-associated t-cell epitopes for vaccine development. Nat Rev Cancer 2, 514 (2002). https://doi.org/10.1038/nrc841
Issue Date:
DOI: https://doi.org/10.1038/nrc841