Abstract
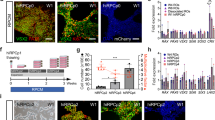
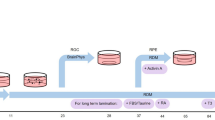
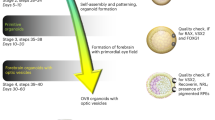
Postmitotic differentiated neurons are among the most difficult cells to reprogram into induced pluripotent stem cells (iPSCs) because they have poor viability when cultured as dissociated cells. To overcome this, other protocols have required the inactivation of the p53 tumor suppressor to reprogram postmitotic neurons, which can result in tumorigenesis of the cells. We describe a method that does not require p53 inactivation but induces reprogramming in retinal cells from reprogrammable mice grown in aggregates with wild-type mouse retinal cells. After the first 10 d of reprogramming, the aggregates are then dispersed and plated on irradiated feeder cells to propagate and isolate individual iPSC clones. The reprogramming efficiency of different neuronal populations at any stage of development can be quantified using this protocol. Reprogramming retinal neurons using this protocol will take 56 d, and these retina-derived iPSCs can undergo retinal differentiation to produce retinae in 34 d. In addition, we describe a quantitative assessment of retinal differentiation from these neuron-derived iPSCs called STEM-RET. The procedure quantifies eye field specification, optic cup formation and retinal differentiation in 3D cultures using molecular, cellular and morphological criteria. An advanced level of cell culture experience is required to carry out this protocol.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Schwartz, S.D. et al. Human embryonic stem cell-derived retinal pigment epithelium in patients with age-related macular degeneration and Stargardt's macular dystrophy: follow-up of two open-label phase 1/2 studies. Lancet 385, 509–516 (2015).
Lamba, D.A., Gust, J. & Reh, T.A. Transplantation of human embryonic stem cell-derived photoreceptors restores some visual function in Crx-deficient mice. Cell Stem Cell 4, 73–79 (2009).
MacLaren, R.E. et al. Retinal repair by transplantation of photoreceptor precursors. Nature 444, 203–207 (2006).
Pearson, R.A. et al. Restoration of vision after transplantation of photoreceptors. Nature 485, 99–103 (2012).
Eiraku, M. & Sasai, Y. Mouse embryonic stem cell culture for generation of three-dimensional retinal and cortical tissues. Nat. Protoc. 7, 69–79 (2012).
Eiraku, M. et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51–56 (2011).
Nakano, T. et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 10, 771–785 (2012).
Hiler, D. et al. Quantification of retinogenesis in 3D cultures reveals epigenetic memory and higher efficiency in iPSCs derived from rod photoreceptors. Cell Stem Cell 17, 101–115 (2015).
Kim, K. et al. Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290 (2010).
Kim, J. et al. Reprogramming of postnatal neurons into induced pluripotent stem cells by defined factors. Stem Cells 29, 992–1000 (2011).
Stadtfeld, M., Maherali, N., Borkent, M. & Hochedlinger, K. A reprogrammable mouse strain from gene-targeted embryonic stem cells. Nat. Methods 7, 53–55 (2010).
Akimoto, M. et al. Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc. Natl. Acad. Sci. USA 103, 3890–3895 (2006).
Lakowski, J. et al. Effective transplantation of photoreceptor precursor cells selected via cell surface antigen expression. Stem Cells 29, 1391–1404 (2011).
Institute of Laboratory Animal Resources. in Guide for the Care and Use of Laboratory Animals (National Research Council, 1996).
Conner, D.A Mouse embryo fibroblast (MEF) feeder cell preparation. Curr. Protoc. Mol. Biol. Chapter 23 Unit 23 22 (2001).
Pruszak, J., Sonntag, K.C., Aung, M.H., Sanchez-Pernaute, R. & Isacson, O. Markers and methods for cell sorting of human embryonic stem cell-derived neural cell populations. Stem Cells 25, 2257–2268 (2007).
Zhang, W.Y, de Almeida, P.E. & Wu, J.C. in StemBook http://www.ncbi.nlm.nih.gov/pubmed/?term=23658974 (2008).
Basu, S., Campbell, H.M., Dittel, B.N. & Ray, A. Purification of specific cell population by fluorescence activated cell sorting (FACS). J. Vis. Exp. http://dx.doi.org/10.3791/1546 (2010).
Acknowledgements
We thank St. Jude Shared Resource Facilities: Flow Cytometry, Light Microscopy, Electon Microscopy, Veterinary Pathology, Cytogenetics and Biomedical Communications for their contributions to data collection and analysis. This work was supported, in part, by a Cancer Center Support grant (CA21765 to M.A.D.) from the National Cancer Institute (NCI), grants to M.A.D. from the NIH (EY014867, EY018599 and CA168875) and the American Lebanese Syrian Associated Charities (ALSAC). M.A.D. was also supported by a grant from Alex's Lemonade Stand Foundation for Childhood Cancer. R.N.E. was supported by grant RO1CA20525 from the NCI. P.A.C. was supported by an NCI training grant (T32CA009657).
Author information
Authors and Affiliations
Contributions
D.J.H. and M.A.D. developed the concepts, designed and performed experiments, analyzed data and developed the STEM-RET analysis. D.J.H. and M.E.B. wrote the paper, designed figures and performed data analysis. M.A.D. and L.M.G. supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Hiler, D., Barabas, M., Griffiths, L. et al. Reprogramming of mouse retinal neurons and standardized quantification of their differentiation in 3D retinal cultures. Nat Protoc 11, 1955–1976 (2016). https://doi.org/10.1038/nprot.2016.109
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2016.109
This article is cited by
-
Hydrogel-based milliwell arrays for standardized and scalable retinal organoid cultures
Scientific Reports (2020)
-
Stem cells and genome editing: approaches to tissue regeneration and regenerative medicine
Journal of Human Genetics (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.