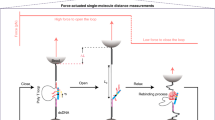
Abstract
Using the scanning probe technique known as Kelvin probe force microscopy it is possible to successfully devise a sensor for charged biomolecules. The Kelvin probe force microscope is a tool for measuring local variations in surface potential across a substrate of interest. Because many biological molecules have a native state that includes the presence of charge centres (such as the negatively charged backbone of DNA), the formation of highly specific complexes between biomolecules will often be accompanied by local changes in charge density. By spatially resolving this variation in surface potential it is possible to measure the presence of a specific bound target biomolecule on a surface without the aid of special chemistries or any form of labelling. The Kelvin probe force microscope presented here is based on an atomic force microscopy nanoprobe offering high resolution (<10 nm), sensitivity (<50 nM) and speed (>1,100 µm s−1), and the ability to resolve as few as three nucleotide mismatches.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Nonnenmacher, M., O'Boyle, M. P. & Wickramsinghe, H. K. Kelvin probe force microscopy. Appl. Phys. Lett. 58, 2921–2923 (1991).
Fodor, S. P. A. et al. Multiplexed biochemical assays with biological chips. Nature 364, 555–556 (1993).
Schena, M., Shalon, D., Davis, R. W. & Brown, P. O. Quantitative monitoring of gene-expression patterns with a complementary-DNA microarray. Science 270, 467–470 (1995).
Drmanac, R. et al. DNA-sequence determination by hybridization—a strategy for efficient large-scale sequencing. Science 260, 1649–1653 (1993).
Schena, M. et al. Microarrays: biotechnology's discovery platform for functional genomics. Trends Biotechnol. 16, 301–306 (1998).
Ramsay, G. DNA chips: State-of-the-art. Nature Biotechnol. 16, 40–44 (1998).
Dufva, M. Fabrication of high quality microarrays. Biomol. Eng. 22, 173–184 (2005).
Ekins, R., Chu, F. & Biggart, E. Development of microspot multi-analyte ratiometric immunoassay using dual fluorescent-labeled antibodies. Anal. Chim. Acta 227, 73–96 (1989).
Ekins, R. & Chu, F. Multianalyte microspot immunoassay—the microanalytical compact-disk of the future. Ann. Biol. Clin.-Paris 50, 337–353 (1992).
Ekins, R. P. & Chu, F. W. Multianalyte microspot immunoassay—microanalytical compact-disk of the future. Clin. Chem. 37, 1955–1967 (1991).
Frank, R. Spot-synthesis—an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron 48, 9217–9232 (1992).
Ekins, R. P. & Chu, F. W. Miniaturized microspot multianalyte immunoassay systems. ACS Symp. Ser. 586, 153–174 (1995).
Fodor, S. P. A. et al. Light-directed, spatially addressable parallel chemical synthesis. Science 251, 767–773 (1991).
Thompson, M. & Cheran, L. E. Surface immobilized biochemical macromolecules studied by scanning Kelvin microprobe. Faraday Discuss. 116, 23–34 (2000).
Cheran, L. E., Sadeghi, S. & Thompson, M. Scanning Kelvin nanoprobe detection in materials science and biochemical analysis. Analyst 130, 1569–1576 (2005).
Cheran, L. E., Chacko, M., Zhang, M. Q. & Thompson, M. Protein microarray scanning in label-free format by Kelvin nanoprobe. Analyst 129, 161–168 (2004).
Thompson, M. et al. Label-free detection of nucleic acid and protein microarrays by scanning Kelvin nanoprobe. Biosens. Bioelectron. 20, 1471–1481 (2005).
Hansen, D. C., Hansen, K. M., Ferrell, T. L. & Thundat, T. Discerning biomolecular interactions using Kelvin probe technology. Langmuir 19, 7514–7520 (2003).
Laoudj, D., Guasch, C., Renault, E., Bennes, R. & Bonnet, J. Surface potential mapping of dispersed proteins. Anal. Bioanal. Chem. 381, 1476–1479 (2005).
Demers, L. M. et al. Direct patterning of modified oligonucleotides on metals and insulators by dip-pen nanolithography. Science 296, 1836–1838 (2002).
Jacobs, H. O., Leuchtmann, P., Homan, O. J. & Stemmer, A. Resolution and contrast in Kelvin probe force microscopy. J. Appl. Phys. 84, 1168–1173 (1998).
Belaidi, S., Girard, P. & Leveque, G. Electrostatic forces acting on the tip in atomic force microscopy: Modelization and comparison with analytic expressions. J. Appl. Phys. 81, 1023–1030 (1997).
Vettiger, P. et al. The ‘Millipede’—more than one thousand tips for future AFM data storage. IBM J. Res. Dev. 44, 323–340 (2000).
Hansma, P. K., Schitter, G., Fantner, G. E. & Prater, C. Applied physics — high-speed atomic force microscopy. Science 314, 601–602 (2006).
Lord Kelvin . Contact electricity of metals. Philos. Mag. 46, 82–120 (1898).
Zisman, W. A. A new method of measuring contact potential differences in metals. Rev. Sci. Instrum. 3, 367–368 (1932).
Weaver, J. M. R. & Abraham, D. W. High-resolution atomic force microscopy potentiometry. J. Vac. Sci. Technol. B 9, 1559–1561 (1991).
Acknowledgements
This research was performed under an appointment to the Department of Homeland Security (DHS) Scholarship and Fellowship Program (A.K.S.), administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the US Department of Energy (DOE) and DHS. ORISE is managed by Oak Ridge Associated Universities (ORAU) under DOE contract number DE-AC05-06OR23100. All opinions expressed in this paper are those of the authors and do not necessarily reflect the policies and views of DHS, DOE or ORAU/ORISE. This research was funded through the Packard Fellows Program (A.M.B.) from the David and Lucile Packard Foundation.
Author information
Authors and Affiliations
Contributions
A.K.S. and A.M.B. conceived the experiments. A.K.S. designed and performed the experiments. A.K.S. and A.M.B. wrote the manuscript. Both authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary figures S1–S5 (PDF 483 kb)
Rights and permissions
About this article
Cite this article
Sinensky, A., Belcher, A. Label-free and high-resolution protein/DNA nanoarray analysis using Kelvin probe force microscopy. Nature Nanotech 2, 653–659 (2007). https://doi.org/10.1038/nnano.2007.293
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nnano.2007.293
This article is cited by
-
Electrostatic Read Out for Label-Free Assays Based on Kelvin Force Principle
Sensing and Imaging (2019)
-
Evaluation of surface charge shift of collagen fibrils exposed to glutaraldehyde
Scientific Reports (2018)
-
Nanoelectrical characterization of amyloid-β42 aggregates via Kelvin probe force microscopy
Macromolecular Research (2017)
-
Enhanced electrostatic force microscopy reveals higher-order DNA looping mediated by the telomeric protein TRF2
Scientific Reports (2016)
-
Routing of individual polymers in designed patterns
Nature Nanotechnology (2015)