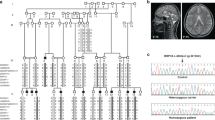
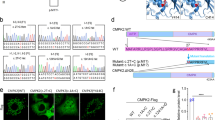
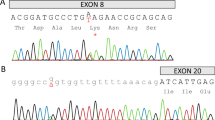
Abstract
Although ribosomes are ubiquitous and essential for life, recent data indicate that monogenic causes of ribosomal dysfunction can confer a remarkable degree of specificity in terms of human disease phenotype. Box C/D small nucleolar RNAs (snoRNAs) are evolutionarily conserved non-protein-coding RNAs involved in ribosome biogenesis. Here we show that biallelic mutations in the gene SNORD118, encoding the box C/D snoRNA U8, cause the cerebral microangiopathy leukoencephalopathy with calcifications and cysts (LCC), presenting at any age from early childhood to late adulthood. These mutations affect U8 expression, processing and protein binding and thus implicate U8 as essential in cerebral vascular homeostasis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Change history
19 September 2016
In the version of this article initially published, the names of authors Geraldine Aubert, Gerardine Quaghebeur and Yoann Rose were misspelled. The error has been corrected in the print, HTML and PDF versions of the article.
References
Labrune, P. et al. Extensive brain calcifications, leukodystrophy, and formation of parenchymal cysts: a new progressive disorder due to diffuse cerebral microangiopathy. Neurology 46, 1297–1301 (1996).
Nagae-Poetscher, L.M. et al. Leukoencephalopathy, cerebral calcifications, and cysts: new observations. Neurology 62, 1206–1209 (2004).
Corboy, J.R., Gault, J. & Kleinschmidt-DeMasters, B.K. An adult case of leukoencephalopathy with intracranial calcifications and cysts. Neurology 67, 1890–1892 (2006).
Livingston, J.H. et al. Leukoencephalopathy with calcifications and cysts: a purely neurological disorder distinct from coats plus. Neuropediatrics 45, 175–182 (2014).
Watkins, N.J. & Bohnsack, M.T. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA 3, 397–414 (2012).
Peculis, B.A., DeGregorio, S. & McDowell, K. The U8 snoRNA gene family: identification and characterization of distinct, functional U8 genes in Xenopus. Gene 274, 83–92 (2001).
Kiss, T., Fayet, E., Jády, B.E., Richard, P. & Weber, M. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb. Symp. Quant. Biol. 71, 407–417 (2006).
Tomasevic, N. & Peculis, B.A. Xenopus LSm proteins bind U8 snoRNA via an internal evolutionarily conserved octamer sequence. Mol. Cell. Biol. 22, 4101–4112 (2002).
Kufel, J., Allmang, C., Petfalski, E., Beggs, J. & Tollervey, D. Lsm proteins are required for normal processing and stability of ribosomal RNAs. J. Biol. Chem. 278, 2147–2156 (2003).
Watkins, N.J., Lemm, I. & Lührmann, R. Involvement of nuclear import and export factors in U8 box C/D snoRNP biogenesis. Mol. Cell. Biol. 27, 7018–7027 (2007).
Watkins, N.J. et al. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103, 457–466 (2000).
Watkins, N.J., Dickmanns, A. & Lührmann, R. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 22, 8342–8352 (2002).
Anderson, B.H. et al. Mutations in CTC1, encoding conserved telomere maintenance component 1, cause Coats plus. Nat. Genet. 44, 338–342 (2012).
Takai, H. et al. A POT1 mutation implicates defective telomere end fill-in and telomere truncations in Coats plus. Genes Dev. 30, 812–826 (2016).
Briggs, T.A. et al. Cerebroretinal microangiopathy with calcifications and cysts (CRMCC). Am. J. Med. Genet. A. 146A, 182–190 (2008).
Polvi, A. et al. Mutations in CTC1, encoding the CTS telomere maintenance complex component 1, cause cerebroretinal microangiopathy with calcifications and cysts. Am. J. Hum. Genet. 90, 540–549 (2012).
Christopher, K.J., Wang, B., Kong, Y. & Weatherbee, S.D. Forward genetics uncovers transmembrane protein 107 as a novel factor required for ciliogenesis and Sonic hedgehog signaling. Dev. Biol. 368, 382–392 (2012).
Shaheen, R. et al. Identification of a novel MKS locus defined by TMEM107 mutation. Hum. Mol. Genet. 24, 5211–5218 (2015).
Hanks, S. et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat. Genet. 36, 1159–1161 (2004).
Scheper, G.C. et al. Mitochondrial aspartyl-tRNA synthetase deficiency causes leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation. Nat. Genet. 39, 534–539 (2007).
Lafontaine, D.L. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat. Struct. Mol. Biol. 22, 11–19 (2015).
Tyc, K. & Steitz, J.A. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 8, 3113–3119 (1989).
Peculis, B.A. & Steitz, J.A. Sequence and structural elements critical for U8 snRNP function in Xenopus oocytes are evolutionarily conserved. Genes Dev. 8, 2241–2255 (1994).
Peculis, B.A. The sequence of the 5′ end of the U8 small nucleolar RNA is critical for 5.8S and 28S rRNA maturation. Mol. Cell. Biol. 17, 3702–3713 (1997).
Freed, E.F., Bleichert, F., Dutca, L.M. & Baserga, S.J. When ribosomes go bad: diseases of ribosome biogenesis. Mol. Biosyst. 6, 481–493 (2010).
Ramagopal, S. Are eukaryotic ribosomes heterogeneous? Affirmations on the horizon. Biochem. Cell Biol. 70, 269–272 (1992).
Xue, S. & Barna, M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 13, 355–369 (2012).
Abecasis, G.R., Cherny, S.S., Cookson, W.O. & Cardon, L.R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 30, 97–101 (2002).
Kong, A. & Cox, N.J. Allele-sharing models: LOD scores and accurate linkage tests. Am. J. Hum. Genet. 61, 1179–1188 (1997).
Dobbyn, H.C. & O'Keefe, R.T. Analysis of Snu13p mutations reveals differential interactions with the U4 snRNA and U3 snoRNA. RNA 10, 308–320 (2004).
Acknowledgements
We are very grateful to the affected families for their involvement in our research. The authors would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. DNA panels from the NINDS Human Genetics Resource Center DNA and Cell Line Repository were used in this study, as well as clinical data. The submitters who contributed samples are acknowledged in detailed descriptions of each panel: NDPT099 and NDPT095. Y.J.C. acknowledges funding from the Newlife Foundation (14-15/15), the Great Ormond Street Hospital Children's Charity (V1212) and a state subsidy managed by the National Research Agency (France) under 'Investments for the Future' (ANR-10-IAHU-01). R.T.O'K. acknowledges the Wellcome Trust (104981). This work was supported by Wellcome Trust funding (097820/Z/11/B) to Y.J.C. and R.T.O'K. P.R. has received research support from La Ligue (Equipe Lab Elisée) and the Centre National de la Recherche Scientifique (CNRS). This paper is dedicated to the memory of John L. Tolmie.
Author information
Authors and Affiliations
Contributions
Exome sequencing was performed by J.E.U., J.O'S., S.G.W. and S.S.B. Exome and genomic capture data were analyzed by E.M.J. Linkage analysis was undertaken by J.E.U. Sanger sequencing and cloning were performed by E.M.J. with assistance from A.O. and L.C.G. Copy number analysis and microsatellite genotyping were undertaken by E.M.J. Cell lines were maintained by E.M.J., A.O., L.C.G., M.P.R. and Y.R. In vitro transcription of U8 snoRNA, EMSAs, 3′-end processing assays, luciferase assays and polysome assays were performed by E.M.J., C.J.K. and R.T.O'K. with assistance from G.D.P. Cell proliferation, senescence and apoptosis assays were performed by M.P.R. DNA content and ImmuoFISH assays were undertaken by M.P.R. with assistance from P.R. Immunoblotting was performed by P.R.K. RT–PCR was performed by M.P.R. with assistance from G.I.R. RNA modeling studies were performed by S.G.-J. Telomere analysis was undertaken by G.M.B., M. Haubitz and G.A. Y.J.C. and R.T.O'K. designed and supervised the project and wrote the manuscript supported by G.I.R. and E.M.J. M.S.v.d.K., J.H.L. and Y.J.C. reviewed the patient scans. K.W.B., A.J.B., R.B., A.B., J.E.B.-H., J.A.B., D.M. Cassiman, R.C., D.M. Cordelli, L.M.D.W., A.J.F., P.F., N.A.F., A.E.F., H.G., C.A.H., I.H., R.J.J., R.K., G.Q., L.L., C.M.L., T.J.M., S.G.M., I.M., S.N., K.Õ., P.P., R.S., E.H.S., C.S., H.S.S., J.S., C.U., H.V.E., C.E.G.V.M., A.V., E.L.W., E.M.B., P.G.L., A.P., K.R.S., M. Haubitz, M. Henneke and A.W. identified affected patients or assisted with related clinical and laboratory studies.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Histopathological characteristics of LCC.
(a,b) Cerebellar brain biopsy taken from F1172 at the age of 50 years showing numerous blood vessels, with groups of vessels that resemble angioma. (c,d) The vessels appear ectatic, partially thickened and hyalinized, and demonstrate an irregular vessel wall. Inflammatory changes are not evident. (d–f) Numerous macrophages with hemosiderin deposits around vessels (arrows) (d,e) and diffusely within the tissue (f) indicate old hemorrhage. (g,h) Both vascular (g) and parenchymal (h) mineralization is seen. (i,j) An extensive gliosis with strikingly high numbers of Rosenthal fibers is present in the parenchyma (some indicated by arrows) (i) and around vessels (arrows) (j). Staining: hematoxylin and eosin in a, c and e–j, Elastica van Gieson in b and d. Scale bars: 200 μm in a–c and f, 100 μm in d, e, g and j, 500 μm in h, and 50 μm in i.
Supplementary Figure 2 Linkage analysis in seven families.
Nonparametric linkage analysis was performed using the Merlin package in five pairs of affected siblings (F331, F426, F454, F521 and F780) born to unrelated parents and two singletons (F344 and F446) who were the product of independent consanguineous unions. K&C lin, Kong and Cox linear model; K&C exp, Kong and Cox exponential model.
Supplementary Figure 3 SNORD118 copy number analysis in F819.
Copy number analysis was performed in F819 using DNA from the two affected individuals (F819_1 and F819_2) and their mother (F819_M; paternal DNA was not available). These data indicate loss of the paternally derived allele in the affected individuals. The parents from three other LCC families were used as comparators, with F619_M chosen as the calibrator sample.
Supplementary Figure 4 Multiple-sequence alignment of SNORD118 sequences.
SNORD118 homologs were identified using Ensembl and Rfam and aligned using CLUSTALW. The alignment was manually refined using RALEE. Nucleotide alterations (not including deletions, insertions and duplications) identified in patients with LCC are annotated above the sequence. From 5′ to 3′, the C box, LSm-binding sites and D box are highlighted by red lines. Aligned columns are colored according to sequence conservation: black, >80% identity; dark gray, >60% identity; light gray, >40% identity. A possible base-paired secondary structure, based on the consensus U8 structure in the Rfam database and analysis with RNAalifold, is shown in dot-bracket notation below the alignment. The following sequences were used for alignment: Homo sapiens, ENST00000363593; Pan troglodytes, ENSPTRT00000052278; Macaca mulatta, ENSMMUT00000034372; Bos taurus, ENSBTAT00000060500; Canis familiaris, ENSCAFT00000034685; Mus musculus, ENSMUST00000082965; Oryctolagus cuniculus, ENSOCUT00000018667; Gallus gallus, ENSGALT00000043652; Xenopus tropicalis, ENSXETT00000065858; Tetraodon nigroviridis, ENSTNIT00000023953; Danio rerio, ENSDART00000115749.
Supplementary Figure 5 In silico analysis of variants in patients with LCC.
In silico analysis of variants located in the stem of a highly conserved hairpin loop from the predicted structure of U8. Bases 96–124 of the wild-type sequence of U8 are shown with patient variants underneath. The predicted base-paired secondary structure of each sequence is shown in dot-bracket notation, together with the minimum folding free energy (kcal/mol). Each of the three variants detected in patients with LCC is predicted to decrease the stability of the hairpin.
Supplementary Figure 6 15.5K–U8 snoRNA EMSA control experiments.
(a) Addition of an antibody to 6×His (ab18184) causes a supershift, demonstrating that the observed shift in U8 RNA is a result of binding to His-15.5K. (b) A competition assay to demonstrate binding specificity. An excess of unlabeled in vitro–transcribed wild-type U8 RNA (2.5 μg) is sufficient to outcompete 5′-end-labeled in vitro–transcribed U8 RNA and eliminate the band shift. (c) Coomassie-stained gel showing purified His-15.5K protein. Lane 1, Precision Plus Protein Dual-Color Standard (Bio-Rad); lane 2, purified His-15.5K protein.
Supplementary Figure 7 U8 snoRNA precursor processing control experiments.
(a) 3′-end precursor processing of precursor U8 variants in the absence of RNasin. Removal of commercially available RNase inhibitors (RNasin, Promega) does not alter the in vitro 3′-end processing of 5′-end-labeled precursor U8 snoRNA (U8-165) in HeLa nuclear extracts. (b) 3′-end precursor processing of 5′-end-labeled precursor U8 variant n.58A>G. At 30 min, the pattern of processing intermediates is the same as for wild-type RNA.
Supplementary Figure 8 Cell cycle analysis.
Comparison of the effect of mitomycin C treatment on cell cycling in primary fibroblasts from a patient with Fanconi anemia (n = 1) (positive control), patients with LCC (n = 3) and healthy controls (n = 3). Top, representative histogram of DNA content in untreated and mitomycin C–treated cells. Bottom left, bar graph representation of the percentage of treated or untreated cells in G0/1, S or G2/M phase. Data shown are means ± s.e.m. Bottom right, to be able to compare the effect of mitomycin C on cell cycling between controls and patients with LCC, as compared to cells from a patient with Fanconi anemia, we calculate a mitomycin score with the following formula: (% increase in S + G2/M phase fraction after treatment/% increase in S + G2/M phase fraction of Fanconi cells after treatment) × 100. No significant difference were detected by Mann–Whitney U testing; n = 2 experiments.
Supplementary Figure 9 Polysome analysis in lymphoblast cell lines.
Polysome profiles in extracts of EBV-transformed LCLs. Extracts from two patient lines were compared to a healthy control and did not demonstrate consistent abnormalities in translation efficiency.
Supplementary Figure 10 Comparison of neuroimaging in patients with CP due to CTC1 mutations and LCC due to mutations in SNORD118 (F172).
(a,c) Axial T2 MRIs of a patient with CP (a) and a patient with LCC (c) showing a similar appearance with leukoencephalopathy, calcification and cysts. (b,d) CT images are also largely similar with dense thalamic and deep cortical calcification. There is more extensive calcification in the patient with LCC (d).
Supplementary Figure 11 CTC1 expression in patients with LCC and controls.
qRT–PCR of CTC1 expression in primary fibroblast cell lines from three controls and four patients with LCC, normalized to two housekeeping genes, HPRT1 and 18S. RQ is equal to 2− ΔΔCt, that is, the normalized fold change relative to a control.
Supplementary Figure 12 53BP1 and telomere-dysfunction-induced foci.
(a) Representative images of 53BP1 staining of control, LCC and CTC1 primary fibroblasts. Scale bar, 2.5 μm. (b) Comparison of the number of 53BP1 foci in primary fibroblasts from healthy controls (n = 3), patients with LCC (n = 4) and patients with CTC1 (n = 3). The red bar represents the median value for each group. n = 2 independent experiments. (c) Representative images of a TIF are displayed for a control, LCC and CTC1 cell line. Scale bar, 2.5 μm. (d) Comparison of the number of TIFs in primary fibroblasts from patients with CTC1 (n = 3), patients with LCC (n = 4) and healthy controls (n = 2). The red bar represents the median value for each group. Data were derived from n = 3 independent experiments and are grouped. Kruskal–Wallis with Dunn’s multiple-comparison test: *P < 0.05, ** P < 0.01. No significant difference between patients with LCC and controls was detected. Black circle, CTRL1; black square, CTRL2; black triangle, CTRL3; open diamond, F281; open circle, F334; open square, F691; open triangle, F906; gray triangle, F345; gray circle, F1314_1; gray square, F1314_2.
Supplementary Figure 13 TMEM107/TMEM107 expression (RNA/protein).
(a) qRT–PCR of TMEM107 expression in primary fibroblast cell lines from three controls and four patients with LCC, normalized to two housekeeping genes, HPRT1 and 18S. RQ is equal to 2− ΔΔCt, that is, the normalized fold change relative to a control. (b) Immunoblot of TMEM107 in primary fibroblast cell lines from three controls and five patients with LCC. Whole-cell lysates were derived from 5 × 106 cells per sample, and 30 μg of total protein was loaded per lane. Immunoblotting for actin (42 kDa) was used as a loading control.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–13 and Supplementary Tables 1–8. (PDF 6081 kb)
Rights and permissions
About this article
Cite this article
Jenkinson, E., Rodero, M., Kasher, P. et al. Mutations in SNORD118 cause the cerebral microangiopathy leukoencephalopathy with calcifications and cysts. Nat Genet 48, 1185–1192 (2016). https://doi.org/10.1038/ng.3661
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3661
This article is cited by
-
Clinical and neuroimaging review of monogenic cerebral small vessel disease from the prenatal to adolescent developmental stage
Japanese Journal of Radiology (2024)
-
Leukoencephalopathy with calcifications and cysts: A case report with literature review
Neurological Sciences (2023)
-
Leukoencephalopathy with brain calcifications and cysts (Labrune syndrome) case report: diagnosis and management of a rare neurological disease
BMC Neurology (2022)
-
Paediatric neurosurgical implications of a ribosomopathy: illustrative case and literature review
Child's Nervous System (2022)
-
Optimized photochemistry enables efficient analysis of dynamic RNA structuromes and interactomes in genetic and infectious diseases
Nature Communications (2021)