Abstract
Methane is an essential component of the global carbon cycle and one of the most powerful greenhouse gases, yet it is also a promising alternative source of carbon for the biological production of value-added chemicals. Aerobic methane-consuming bacteria (methanotrophs) represent a potential biological platform for methane-based biocatalysis. Here we use a multi-pronged systems-level approach to reassess the metabolic functions for methane utilization in a promising bacterial biocatalyst. We demonstrate that methane assimilation is coupled with a highly efficient pyrophosphate-mediated glycolytic pathway, which under oxygen limitation participates in a novel form of fermentation-based methanotrophy. This surprising discovery suggests a novel mode of methane utilization in oxygen-limited environments, and opens new opportunities for a modular approach towards producing a variety of excreted chemical products using methane as a feedstock.
Similar content being viewed by others
Introduction
Nature provides two alternative forms of methane as a resource: natural gas, relatively abundant today but still a non-renewable fossil fuel, and renewable biogas, a byproduct of modern society that is often wasted1,2,3. Interest in new technologies for effective conversion of flared/waste sources of methane into chemical compounds, including next-generation fuels, continues to increase4,5,6. The use of microbial cells and enzymes as catalysts for methane conversion represents an appealing approach in this context7,8,9,10,11. The benefits of methane biotechnology include a self-sustainable component, as any biomass generated could be used as single cell protein or converted back to methane via anaerobic digestion. However, besides single cell protein and polyhydroxybutyrate, exploitation of methane-based catalysis for the production of chemicals and fuels has not yet proven successful at the commercial level.
Gammaproteobacterial methanotrophs with the ribulose monophosphate (RuMP) pathway are among the most promising microbial systems for methane-based biotechnology. Reconstruction of the methane utilization network in these methanotrophs has been based on a number of biochemical studies that pointed towards the Entner–Doudoroff (EDD)-variant of the RuMP pathway as the major route for single carbon (C1) assimilation12,13,14. The major biochemical evidence that favoured the EDD variant of the RuMP cycle included relatively high activities of two key enzymes of the pathway (6-phosphogluconate dehydratase and 2-keto-3-deoxy-6-phosphogluconate aldolase) and multiple enzymatic lesions in the Embden–Meyerhof–Parnas (EMP) pathway15,16. Activity of pyruvate kinase has not been detected previously in any gammaproteobacterial methanotroph16,17. The presence of a reversible pyrophosphate (PPi)-dependent phosphofructotransferase led to the conclusion that the EMP pathway represents a metabolic loop balancing the level of glyceraldehyde-3-phosphate and phosphoenolpyruvate (PEP)16,18. This metabolic arrangement has served as the foundation for theoretical characterization of efficiency and yield of methane utilization14,15. However, the predicted maximum carbon conversion efficiency (39–47%) was considerably less than measured values (64–66.5%)15,19. Theoretically, the RuMP pathway can provide a better efficiency of carbon utilization when operated via the EMP pathway than via the EDD pathway14. Genomic studies indicate that a complete set of EMP pathway enzymes is encoded in the genomes of all sequenced methanotrophic bacteria harbouring the RuMP pathway for methane assimilation. Furthermore, proteomic studies showed that both pathways are expressed in Methylococcus capsulatus20.
In this work, we renew the current understanding of metabolic functions essential for methane utilization through detailed investigations of C1-assimilation in Methylomicrobium alcaliphilum strain 20Z, a haloalkalitolerant methanotroph that is a promising biocatalyst11. Availability of the M. alcaliphilum 20Z genome sequence21 allowed us to apply systems-level approaches including genome-wide transcriptomic studies (Illumina-based RNA-Seq), metabolomics and 13C-label distribution analysis of methane-grown cultures for metabolic reconstruction of C1 utilization pathways in this strain.
Results
Transcriptomic study
M. alcaliphilum 20Z grown aerobically with methane as the sole source of carbon and energy showed high levels of expression for genes known to be involved in the metabolism of C1 compounds including those for membrane-bound methane monooxygenase (pmoCAB), PQQ-dependent methanol dehydrogenase (mxaFIG) and two key enzymes of the RuMP pathway-hexulose phosphate synthase (hps) and phosphohexuloisomerase (hpi) (Fig. 1a; Supplementary Table S1), as expected. The relative abundance of transcripts for enzymes involved in the RuMP pathway downstream from fructose-6-phosphate were 3–20-fold lower than those of hpi and hps. Remarkably the abundance of transcripts encoding glycolytic pathway enzymes was 2–10-fold higher than EDD pathway enzymes (that is, edd and eda) (Supplementary Table S1). Furthermore, one of the putative pyruvate kinase genes, pyk2 (MALCv4_3080), showed high expression.
(a) Predicted positions of incorporated 13C (indicted in red) for the EDD (dark red arrows) and the EMP (blue arrows) variants. Dashed lines indicate multistep reactions. The pentose-phosphate pathway variant for regeneration of ribulose 5-phosphate is indicated by orange dashed arrows. Enzyme activities (in nmol min1 per mg protein) in cell free extracts of M. alcaliphilum 20Z were as follows: methane monoxygenase (Mmo), 70±5; PQQ-dependent methanol dehydrogenase (Mdh), 230±12; NAD-dependent formate dehydrogenase (Fdh), 130±7; hexulose phosphate synthase/hexulose phosphate isomerase (Hps/Hpi), 600±30; glucose phosphate isomerase (Gpi), 32±5; NADP-dependent glucose 6-phosphate dehydrogenase, 34±2 (Gpd); NAD-dependent glucose 6-phosphate dehydrogenase, 23±2 (Gdp); NADP-dependent 6-phosphogluconate dehydrogenase (Edd), 32±2; KDPG aldolase (Eda), 60±4; fructose-bisphosphate aldolase (Fba), 35± 2; PPi-phosphofructokinase (Pfk), 70± 4. (b) Summary equations for production of 3-phosphoglycerate from formaldehyde via the RuMP pathway for the EDD variant (1) or the EMP variant with either the ATP-dependent EMP pathway (2) or the PPi-dependent EMP pathway (3). Abbreviations: Ru5P, ribulose 5-phosphate; He6P, 3-hexulose 6-phosphate; F6P, fructose 6-phosphate; KDPG, 2-keto-3-deoxy 6-phosphogluconate; F1,6P, fructose 1,6-bisphosphate; DAP, dihydroxyacetone phosphate; GAP, glyceraldehyde 3-phosphate; PGA, 3-phosphoglycerate; Pi, inorganic phosphate.
Characterization of methanotrophic pyruvate kinase
In accordance with previous studies, no pyruvate-forming activity was detected in cell-free extracts of M. alcaliphilum strain 20Z with three different enzymatic assays (see Methods). However, when the pyruvate kinase gene was overexpressed in Escherichia coli, purified protein preparations displayed significant pyruvate kinase activity (7 U mg−1 of protein at the optimum pH 7.5) (Table 1 and Supplementary Table S2). We found that the enzyme activity was strongly stimulated (20-fold) in the presence of a set of the RuMP pathway intermediates: glucose-6-phosphate, fructose-6-phosphate, ribose-5-phosphate, ribulose-5-phosphate or erythrose-4-phosphate (Supplementary Table S2). ATP, PPi and Pi strongly inhibited enzyme activity; however, this inhibitory effect was completely abolished by the addition of activators such as fructose-6-phosphate or ribose-5-phosphate. Overall, the data suggest that the strain possesses an active pyruvate kinase that is strongly dependent on the presence of RuMP cycle intermediates. It should be mentioned that the pyruvate kinase does not show any activity in cell extracts from methanotrophic bacteria, even after supplementation with the inducers shown in Table 1 and Supplementary Table S2. As pyk2 is one of the highly expressed genes in the transcriptome of strain 20Z, and the protein is readily detected (Kalyuzhnaya, unpublished data), we can only speculate that in methanotrophs the enzyme is not stable or is inactivated by another yet unknown component, such as a tightly bound inhibitor, a protease or a modifying protein. However, the kinetic properties of Pyk2 indicate that significant flux of cell carbon could occur via the EMP pathway in methanotrophic bacteria. In order to further test this hypothesis, we analysed the intracellular concentrations of pathway intermediates using a metabolomic approach.
EMP pathway is the main route for C1-carbon assimilation
In cells of M. alcaliphilum 20Z grown on methane, the intracellular abundance of the majority of EMP pathway intermediates is high (Supplementary Table S3). In contrast, two key intermediates of the EDD pathway, 6-phosphogluconate and 2-dehydro-3-deoxy-phosphogluconate were only barely detected in cell samples. To further probe the metabolic pathway for methane assimilation in M. alcaliphilum 20Z, we monitored the dynamic incorporation of 13C-labelled methane into downstream intermediates of the RuMP pathway (Supplementary Fig. S1). As expected, the relative abundance of fructose 6-phosphate/glucose 6-phosphate increased at very early time points, demonstrating that methane was rapidly assimilated through the reaction of Ru5P and 13C-labelled formaldehyde. The downstream metabolites phosphoglycerate and PEP were also sequentially labelled. Owing to the low pool size, we were not able to estimate the rate of incorporation of 13C-carbon into intermediates of the EDD pathway.
In order to distinguish between the two pathways, we performed 13C-pyruvate tracing analysis coupled with tandem mass spectrometry. If pyruvate is formed via the EDD pathway, the initial 13C-incorporation should be observed in position 1; in contrast, pyruvate derived from PEP through the EMP pathway should be labelled in position 3 (Fig. 1a). As shown in Fig. 2, only a small fraction of pyruvate was labelled in position 1 during the course of the experiment. The rate of 13C incorporation into position 3 of pyruvate was at least six-fold higher than the rate of incorporation into position 1. These experiments confirmed that the major fraction of cellular pyruvate comes from the EMP pathway during growth of the methanotrophic culture on methane. A similar carbon isotopic distribution in pyruvate was observed for Methylomonas sp. LW13, a typical representative of gammaproteobacterial methanotrophic bacteria (Supplementary Fig. S2).
Methane utilization via fermentation
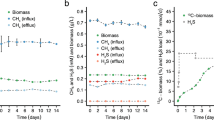
The new arrangement of the methanotrophic network opens up a possibility for fermentation. Methanotrophs require O2 for the oxidation of methane, so experiments were carried out with cells grown in bioreactors in which air was provided at low levels and the dissolved O2 concentrations were kept at undetectable to 0.1%. In a continuous bioreactor culture, M. alcaliphilum 20Z grew slowly, with a doubling time of 23 h. Transcriptomic profiles of batch bioreactor cultures grown at low O2 showed that similar to what is observed for aerobic growth, relative expression of EMP genes is high. The most notable changes in the transcriptome were the downregulation of genes for NADH:ubiquinone oxidoreductase and cytochrome c oxidase, and the upregulation of genes for the O2 carrier bacteriohemerythrin and for the pathways for mixed-acid fermentation and H2 production (Supplementary Table S1 and Supplementary Fig. S3). In our experiments, the expression profile of bacteriohemerythrin, shown to be essential for high in vitro activity of particulate methane monooxygenase in M. capsulatus Bath22, indicated that it most likely contributes to O2-scavenging/partitioning in M. alcaliphilum 20Z. Genes predicted to encode fermentation pathway enzymes with increased expression under microoxic conditions included a putative acetate kinase, 3-ketoacyl-CoA thiolase, 3-hydroxyacyl-CoA dehydrogenase, malate dehydrogenase, fumarase, succinate dehydrogenase and lactate dehydrogenase. Intriguingly, upregulation of genes encoding a NAD-reducing hydrogenase was observed (Supplementary Table S1). These changes suggested production of a set of possible fermentation products, including formate, acetate, succinate, lactate, 3-hydroxybutyrate and H2. Significantly, in bioreactor cultures acetate, succinate, lactate and H2 were detected, but only in medium from batch and chemostat cultures grown at low O2 tension, whereas formate increased about threefold (Table 2). Extracellular concentrations of these acids increased markedly after incubation of low O2 bioreactor samples in closed vials flushed with N2. Low amounts of 3-hydroxybutyrate also accumulated. Furthermore, significant accumulation of H2 was detected in closed vial experiments (Table 2). Similar incubations supplied with 13C-methane confirmed that formic and acetic acids were produced from methane (Supplementary Table S4). The rate of methane consumption in the closed vial experiments was exceptionally low (1.75±0.41 nmol min−1 per mg protein−1); however 3% and 15% of the added methane was consumed in 12 and 60 h, respectively. The total amount of produced extracellular carbon, mostly acetate and formate, was equivalent to 40–50% of the total methane carbon consumed. These data suggest that in the presence of sufficient O2 to drive methane oxidation, M. alcaliphilum 20Z is capable of fermentation from methane-derived formaldehyde, and that methane utilization at low O2 tension involves switching to a novel fermentation mode leading to the formation of formate, acetate, succinate, lactate and hydroxybutyrate as end products, with little biomass synthesis (Fig. 3). The presence of putative fermentation genes in the genome of gammaproteobacterial methanotrophs suggests that this type of metabolism is likely widespread (Supplementary Table S5).
Discussion
The experiments presented here change our understanding of methane assimilation through the RuMP pathway in obligate methanotrophic bacteria in some fundamental aspects. First, utilization of the PPi-mediated EMP pathway significantly increases the predicted efficiency of one-carbon assimilation. Genes encoding a membrane-bound proton-translocating pyrophosphatase show significant expression, suggesting that this enzyme could be one of the possible candidates for the regeneration of PPi from ATP. The predicted ratio of ATP hydrolysis/PPi formation for this class of enzymes is 1:3 (ref. 23). Therefore, not only does the assimilation of 9 mol of formaldehyde by this metabolic scheme to generate three 3-carbon intermediates require no additional energy, it actually produces three moles of reducing power (NADH) and two moles of ATP (Fig. 1b).
Second, the surprising discovery of this ATP-producing assimilatory route raised the possibility of fermentation as a new mode of methane utilization. The ability of methanotrophic cultures to convert methane into excreted organic compounds has previously been described and it has been suggested to support denitrification by wastewater treatment communities19,24,25,26,27. However, metabolic pathways for production of organic compounds from methane were unknown, and excreted compounds were often described as the product of cell lysis or a starvation response24,25,26,27. Our results suggested the possibility that organic acids could be produced as a result of fermentation. This discovery challenges our understanding of methanotrophy as microbial metabolism linked to respiration (O2, NO3− or SO42−) and has major implications for the environmental role of methanotrophic bacteria in removing this greenhouse gas in O2-limited environments. Numerous environmental studies indicate that gammaproteobacterial methanotrophs thrive at oxic–anoxic interfaces28,29,30. If fermentation is the major metabolic mode of methane cycling under O2-limiting conditions, then our understanding of the role of methanotrophic bacteria in supporting the global carbon cycle may be in need of revision in some respects – not so much in terms of carbon being mineralized into CO2, but of the proportion of carbon ending in biomass (Fig. 3). Most notably, in our experiments, only a small fraction of the oxidized methane was converted to biomass, so methanotrophic bacteria may represent only a minor part of the overall microbial community involved in the conversion of methane into biomass. Rather, our results suggest the possibility that in O2-limiting environments, methanotrophs drive the conversion of methane to excreted products and hydrogen, which are then used and transformed by non-methanotrophs. If confirmed by future work, this has implications for microbial community structure and functioning in environments where the methane cycle is prominent.
The surprising discovery of glycolysis-based methane assimilation and production of hydrogen in strain M. alcaliphilum 20Z may open new opportunities to producing a variety of products using methane as a feedstock, provided this pathway may be operated in a sufficiently efficient way in the laboratory. Virtually all biosynthetic modules for the production of a wide variety of chemicals developed for glucose-based catalysis in E. coli could also be implemented in cultures containing this EMP variant of the RuMP pathway. Thus, our demonstration of methane-based fermentation suggests new approaches for the commercial conversion of methane to hydrogen and excreted products using microoxic production conditions.
Methods
Cell cultivation and growth parameters
M. alcaliphilum 20Z cells were grown using a mineral salts medium31 in either closed vials (50 ml culture in 250 ml vials, with shaking at 200 r.p.m.) or bioreactor cultures (fed-batch or chemostat; 1 l working volume in a 2 l bench top BioFlo 110 modular bioreactor, New Brunswick Scientific, Edison, NJ, USA). Cells were grown at 28 oC. Optical density of cell cultures was measured on a Beckman DU 640B spectrophotometer in plastic 1.5 ml cuvettes with a 1 cm path length. Chemostat cultures maintained a steady-state optical density at 600 nm (OD600) of ~2.0±0.2. The dilution rate was 0.12 h−1 for aerobic cultures (influent gas mixture – 20% CH4:20% O2:60 N2, dissolved O2 tension was 49–54%) and 0.03 h−1 for low O2 cultures (influent gas mixture 20% CH4:5% O2:75 %N2; dissolved O2 tension was non-detectable to 0.1%. pH (9.0) was controlled by the automatic addition of 1N NaOH. Agitation was kept constant at 1000 r.p.m. Samples of inflow and outflow gases were collected daily in triplicates for gas analysis. The rates of methane consumption and H2 production were determined by incubating cell samples (50 ml, OD600 between 2 and 4) in closed 250 ml vials for 12–60 h at 28 °C. Before incubation, vials were flushed for 15 min with a gas mixture containing either 20% CH4:5% O2:75% N2 or 20% CH4:80% N2, or 20% 13CH4:80%N2.
Gas analysis
Methane measurements were made on a Shimadzu Gas Chromatograph GC-14A, using an FID detector with helium as the carrier gas. O2 and H2 measurements were made on a multiple gas analyser SRI 8610C Gas Chromatograph equipped with TCD/FID detectors (SRI Instruments). Concentrations of gases were deduced from standard curves.
Gene expression
RNA extraction, sequencing, alignment and mapping were performed as described32.
Metabolite measurement
Metabolic and 13C-labelling studies on batch and fed-batch cultures were carried out as described33 with modification for desalting. Briefly, the dried sample was re-dissolved in 1 ml water and handled according to SPE procedures34. MCX, MAX and WAX cartridges (1 cm3, 30 mg, Waters, Milford, MA, USA) were preconditioned separately. A WAX cartridge was connected beneath a MCX cartridge. Each 1 ml sample was directly loaded through both the MCX and WAX reservoirs. The loaded fraction was collected and made basic with 5% ammonium hydroxide and then loaded into a MAX reservoir, followed by elution. After loading and washing with 1 ml of water, the two adjacent MCX and WAX cartridges were disconnected and eluted separately. All the eluted solutions were dried using a vacuum centrifuge. For liquid chromatography-tandem mass spectrometry analysis, each dried sample was re-dissolved in 50 μl water and pooled. Liquid chromatography-tandem mass spectrometry experiments were carried out on a Waters LC-MS system consisting of a 1,525 μ binary HPLC pump with a 2777C autosampler coupled to a Quattro Micro API triple-quadrupole mass spectrometer (Micromass, Manchester, UK), or a Thermo Scientific TSQ quantum access triple-stage quadrupole mass spectrometer. The HILIC columns (Luna NH2, 250 mm × 2 mm, 5 μm, and ZIC-HILIC, 150 mm × 4.6 mm, 5 μm) employing gradient elution were carried out using the previously described conditions35,36. Sugar phosphates were measured by using an ion pairing-reverse phase method37. Singly labelled pyruvate position was determined by multiple reaction monitoring (MRM) scan mode with an injection volume of 10 μl. The MRM experiments were carried out as described previously38. The dwell time for each MRM transition was 0.08 s. All peaks were integrated using Masslynx Applications Manager (version 4.1) software. Quantification of metabolites was obtained by adding culture-derived global 13C-labelled internal standards before cell extraction37. Relative abundance (%) was obtained by normalizing the pool of each metabolite to the sum of all the targeted metabolites.
Dynamic 13C incorporation
For the 13C methane tracing experiment, M. alcaliphilum 20Z cells grown to mid-exponential phase (OD600=0.6–0.8) on 12C methane in vials or fed-batch bioreactor were rapidly transferred to a fresh flask with the same percentage of 13C methane as the sole carbon source as deduced from a growth curve. At the defined time points, the cell culture was harvested and metabolites were analysed as described above.
Protein purification and characterization
Activities of key enzymes of central metabolic pathways were measured as described18,39,40. Pyruvate-forming activity in cell-free extracts of M. alcaliphilum 20Z was also measured using Pyruvate Assay Kit (BioVision Inc., CA, USA). Recombinant pyruvate kinase PK-ubiqitin-His6 was obtained by cloning of the pyk2 gene (MALCv4_3080) in the vector pHUE and expressing in E. coli BL21 (DE3) cells growing in the presence of 0.5 mM IPTG for 5 h at 37 °C. PK-ubiqitin-His6 enzyme was purified by affinity chromatography on a Ni2+-NTA column as described earlier39.
NMR analysis
To estimate the concentration of metabolites excreted into growth medium, 50 ml samples were collected. Cells were separated by centrifugation (15 min at 2,700 × g), filtration via 0.2 μm filter units followed by ultrafiltration through Amicon@Ultra 3K filters. NMR analyses of the culture media were made using a Bruker AVANCE III 800 MHz or 700 MHz spectrometer equipped with a cryoprobe or a room temperature probe suitable for 1H inverse detection with Z-gradients at 298 K. The solvent, water, was removed from the 1 ml culture media samples by drying the samples using a rotary evaporator. The residue was dissolved in an equal volume of phosphate buffer prepared in deuterated water (0.1 M; pH = 7.4) containing 0.2 mM TSP (3-(trimethylsilyl) propionic-2,2,3,3-d4 acid sodium salt). From this solution, 600 μl was placed in a 5 mm NMR tube for analysis. One-dimensional 1H NMR spectra were obtained using a one pulse sequence that included residual water signal suppression from a pre-saturation pulse during the relaxation delay. For each sample, 32 k data points were acquired using a spectral width of 10,000 Hz and a relaxation delay of 6 s. The data were processed using a spectral size of 32 k points and by multiplying with an exponential window function equivalent to a line broadening of 0.3 Hz. The resulting spectra were phase and baseline corrected and referenced with respect to the internal TSP signal. Metabolite peaks in the spectra were then assigned using chemical shift databases, and the peak areas were obtained by integration. Using these peak areas, along with the known concentration of the internal reference (TSP) and the number of protons each peak represented in the molecule, the metabolite concentrations in the culture media were estimated. Similarly, concentrations for the 13C labelled bacterial products were estimated using 13C satellite peaks of metabolites in the 1H NMR spectra. Bruker Topspin version 3.0 and 3.1 software packages were used for NMR data acquisition and processing, respectively.
Additional information
Accession codes: The RNA-Seq data from this study have been deposited in NCBI’s Gene Expression Omnibus (GEO) database under accession GSE51145.
How to cite this article: Kalyuzhnaya, M.G. et al. Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat. Commun. 4:2785 doi: 10.1038/ncomms3785 (2013).
Accession codes
References
Forster, P. M. & Gregory, J. M. The climate sensitivity and its components diagnosed from Earth radiation budget data. J. Climate 19, 39–52 (2006).
Wuebbler, D. J. & Hayhoe, K. Atmospheric methane and global change. Earth Sci. Rev. 57, 177–210 (2002).
Shindel, D. et al. Simultaneously mitigating near-term climate change and improving human health and food security. Science 335, 183–189 (2012).
Jiang, H. et al. Methanotrophs: multifunctional bacteria with promising applications in environmental bioengineering. Biochem. Eng. J. 49, 277–288 (2010).
Wendlandt, K. D. et al. The potential of methane-oxidizing bacteria for applications in environmental biotechnology. Eng. Life Sci. 10, 87–102 (2010).
Olah, G. A., Goeppert, A. & Prakash, G. K. S. Beyond Oil and Gas: The Methanol Economy Wiley-VCH Verlag Gmb&Co. KGaA, Weiheim (2006).
Podkolzin, S. G., Stangland, E. E., Jones, M. E., Peringer, E. & Lercher, J. A. Methyl chloride production from methane over lanthanum-based catalysts. J. Am. Chem. Soc. 129, 2569–2576 (2007).
Dave, B. Prospects for methanol production. (Eds. Wall J., Harwood C. S., Demain A. L. 235–245ASM Press (2008).
Labinger, J. A. Methane activation in homogeneous systems. Fuel Process. Technol. 4, 325–338 (1995).
Dalton, H. The Leeuwenhoek Lecture 2000 the natural and unnatural history of methane-oxidizing bacteria. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360, 1207–1222 (2005).
Trotsenko, Y. A., Doronina, N. V. & Khmelenina, V. N. Biotechnological potential of aerobic methylotrophic bacteria: a review of current state and future prospects. Appl. Biochem. Microbiol. 41, 433–441 (2005).
Boden, R. et al. Complete genome sequence of the aerobic marine methanotroph Methylomonas methanica MC09. J. Bacteriol. 193, 7001–7002 (2011).
Strom, T., Ferenci, T. & Quayle, J. R. The carbon assimilation pathways of Methylococcus capsulatus, Pseudomonas methanica and Methylosinus trichosporium (OB3b) during growth on methane. Biochemistry 144, 465–476 (1974).
Anthony, C. The Biochemistry of Methylotrophs Academic Press, Inc. Ltd. (1982).
Leak, D. J. & Dalton, H. Growth yields of methanotrophs. Appl. Microbiol. Biot. 23, 477–481 (1986).
Trotsenko, Y. A. & Murrell, J. C. Metabolic aspects of aerobic obligate methanotrophy. Adv. Appl. Microbiol. 63, 183–229 (2008).
Khmelenina, V. N., Kalyuzhnaya, M. G., Starostina, N. G., Suzina, N. E. & Trotsenko, Yu., A. Isolation and characterization of halotolerant alkaliphilic methanotrophic bacteria from Tuva soda lakes. Curr. Microbiol. 35, 257–261 (1997).
Rozova, O. N., Khmelenina, V. N., Vuilleumier, S. & Trotsenko, Y. A. Characterization of recombinant pyrophosphate-dependent 6-phosphofructokinase from halotolerant methanotroph Methylomicrobium alcaliphilum 20Z. Res. Microbiol. 161, 861–868 (2010).
Harwood, J. H. & Pirt, S. J. Quantitative aspects of growth of the methane oxidizing bacterium Methylococcus capsulatus on methane in shake flask and continuous chemostat culture. J. Appl. Bacteriol. 35, 597–607 (1972).
Kao, W. C. et al. Quantitative proteomic analysis of metabolic regulation by copper ions in Methylococcus capsulatus (Bath). J. Biol. Chem. 279, 51554–51560 (2004).
Vuilleumier, S. et al. Genome sequence of the haloalkaliphilic methanotrophic bacterium Methylomicrobium alcaliphilum 20Z. J. Bacteriol. 194, 551–552 (2012).
Chen, K. H.-C. et al. Bacteriohemerythrin bolsters the activity of the particulate methane monooxygenase (pMMO) in Methylococcus capsulatus (Bath). J. Inorg. Biochem. 111, 10–17 (2012).
Scöcke, L. & Schink, B. Membrane-bound proton-translocating pyrophosphatase of Syntrophus gentianae, a syntrophically benzoate-degrading fermenting bacterium. Eur. J. Biochem. 256, 589–594 (1998).
Leak, D. J. & Dalton, H. Growth yields of methanotrophs. I. Effect of copper on the energetics of methane oxidation. Appl. Microbiol. Biot. 23, 470–476 (1986).
Morinaga, Y., Yamanaka, S., Yoshimura, M., Takinami, K. & Hirose, Y. Methane metabolism of the obligate methane-utilizing bacterium Methylomonas flagellate, in methane-limited and oxygen-limited chemostat culture. Agric. Biol. Chem. 43, 2452–2458 (1979).
Rhee, G. Y. & Fuhs, G. W. Wastewater denitrification with one-carbon compounds as energy source. J. Water Pollut. Control Fed. 50, 2111–2119 (1978).
Roslev, P. & King, G. M. Aerobic and anaerobic starvation metabolism in methanotrophic bacteria. Appl. Environ. Microbiol. 61, 1563–1570 (1995).
Auman, A. J., Stolyar, S., Costello, A. M. & Lidstrom, M. E. Molecular characterization of methanotrophic isolates from freshwater lake sediment. Appl. Environ. Microbiol. 66, 5259–5266 (2000).
Reim, A., Luke, C., Krause, S., Pratscher, J. & Frenzel, P. One millimetre makes the difference: high-resolution analysis of methane-oxidizing bacteria and their specific activity at the oxic-anoxic interface in a flooded paddy soil. ISME J 6, 2128–2139 (2012).
Ho, A. et al. Revisiting methanotrophic communities in sewage treatment plants. Appl. Environ. Microbiol. 79, 2841–2846 (2013).
Ojala, D. S., Beck, D. A. C. & Kalyuzhnaya, M. G. Genetic systems for moderately halo(alkali)philic bacteria of the genus Methylomicrobium. Methods Enzymol. 495, 99–118 (2011).
Matsen, J. B., Yang, S., Stein, L. Y., Beck, D. & Kalyuzhnaya, M. G. Global molecular analyses of methane metabolism in methanotrophic Alphaproteobacterium, Methylosinus trichosporium OB3b. Part I. Transcriptomic study. Front. Microbiol. 4, 40 (2013).
Yang, S. et al. Global molecular analyses of methane metabolism in methanotrophic Alphaproteobacterium, Methylosinus trichosporium OB3b. Part II. Metabolomics and 13C-labeling study. Front Microbiol. 4, 70 (2013).
Yang, S., Synovec, R. E., Kalyuzhnaya, M. G. & Lidstrom, M. E. Development of a solid phase extraction protocol coupled with liquid chromatography mass spectrometry to analyze central carbon metabolites in lake sediment microcosms. J. Sep. Sci. 34, 3597–3605 (2011).
Yang, S., Sadilek, M., Synovec, R. E. & Lidstrom, M. E. Liquid chromatography-tandem quadrupole mass spectrometry and comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry measurement of targeted metabolites of Methylobacterium extorquens AM1 grown on two different carbon sources. J. Chromatogr. A 1216, 3280–3289 (2009).
Schiesel, S., Lämmerhofer, M. & Lindner, W. Multitarget quantitative metabolic profiling of hydrophilic metabolites in fermentation broths of β-lactam antibiotics production by HILIC-ESI-MS/MS. Anal. Bioanal. Chem. 396, 1655–1679 (2010).
Buescher, J. M., Moco, S., Sauer, U. & Zamboni, N. Ultrahigh performance liquid chromatography-tandem mass spectrometry method for fast and robust quantification of anionic and aromatic metabolites. Anal. Chem. 82, 4403–4412 (2010).
Yang, S., Sadilek, M. & Lidstrom, M. E. Streamlined pentafluorophenylpropyl column liquid chromatography-tandem quadrupole mass spectrometry and global 13C-labeled internal standards improve performance for quantitative metabolomics in bacteria. J. Chromatogr. A 1217, 7401–7410 (2010).
Reshetnikov, A. S. et al. Characterization of the pyrophosphate-dependent 6-phosphofructokinase from Methylococcus capsulatus Bath. FEMS Microbiol. Lett. 288, 202–210 (2008).
Shishkina, V. N. & Trotsenko, Y. A. Multiple enzymatic lesions in obligate methanotrophic bacteria. FEMS Microbiol. Lett. 13, 237–242 (1982).
Acknowledgements
The work was supported by the National Science Foundation (MCB-0842686), the Department of Energy (DE-SC0005154), CRDF Global (RUB1-2946-PU-09) and the Russian Foundation for Basic Research (RFBR 12-04-32122-a). Support from LABGeM and France Genomique for use of the online comparative genomics analysis platform MicroScope is gratefully acknowledged.
Author information
Authors and Affiliations
Contributions
M.G.K. designed experiments, compiled data and prepared the manuscript; Y.F. and S.Y. performed metabolomics, S.Y. and M.G.K. performed 13C-labelling experiments and analysed the metabolomics data; N.E.S. and M.G.K. isolated RNA for RNAseq experiments; J.C. and A.L. performed bioreactor experiments and measured gas compositions; O.N.R. purified and characterized pyruvate kinase 2 and performed comparative genomics; M.G.K., D.R. and G.A.N.G. performed NMR analyses and analysed the NMR data; D.A.C.B. and M.G.K. analysed RNA-Seq data and performed comparative genomics; F.B., S.V. and Y.A.T. edited the manuscript; V.N.K. and M.E.L. provided conceptual advice and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures S1-S3 and Supplementary Tables S1-S5 (PDF 525 kb)
Rights and permissions
About this article
Cite this article
Kalyuzhnaya, M., Yang, S., Rozova, O. et al. Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat Commun 4, 2785 (2013). https://doi.org/10.1038/ncomms3785
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms3785
This article is cited by
-
A rationally designed miniature of soluble methane monooxygenase enables rapid and high-yield methanol production in Escherichia coli
Nature Communications (2024)
-
Type I methanotrophs dominated methane oxidation and assimilation in rice paddy fields by the consequence of niche differentiation
Biology and Fertility of Soils (2024)
-
The potential of CO2-based production cycles in biotechnology to fight the climate crisis
Nature Communications (2023)
-
Metabolic handoffs between multiple symbionts may benefit the deep-sea bathymodioline mussels
ISME Communications (2023)
-
Phylogenetic and metabolic diversity of microbial communities performing anaerobic ammonium and methane oxidations under different nitrogen loadings
ISME Communications (2023)