Abstract
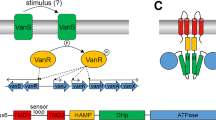
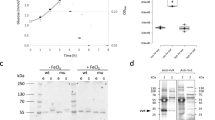
Inducible resistance to the glycopeptide antibiotic vancomycin requires expression of vanH, vanA and vanX, controlled by a two-component regulatory system consisting of a receptor histidine kinase, VanS, and a response regulator, VanR. The identity of the VanS receptor ligand has been debated. Using a synthesized vancomycin photoaffinity probe, we show that vancomycin directly binds Streptomyces coelicolor VanS (VanSsc) and this binding is correlated with resistance and required for vanH, vanA and vanX gene expression.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Leclercq, R., Derlot, E., Duval, J. & Courvalin, P. N. Engl. J. Med. 319, 157–161 (1988).
Courvalin, P. Clin. Infect. Dis. 42 (suppl. 1): S25–S34 (2006).
Pootoolal, J., Neu, J. & Wright, G.D. Annu. Rev. Pharmacol. Toxicol. 42, 381–408 (2002).
Kahne, D., Leimkuhler, C., Lu, W. & Walsh, C. Chem. Rev. 105, 425–448 (2005).
Wright, G.D. Nat. Rev. Microbiol. 5, 175–186 (2007).
Marshall, C.G., Broadhead, G., Leskiw, B. & Wright, G.D. Proc. Natl. Acad. Sci. USA 94, 6480–6483 (1997).
Hong, H.J. et al. Mol. Microbiol. 52, 1107–1121 (2004).
Arthur, M., Molinas, C. & Courvalin, P. J. Bacteriol. 174, 2582–2591 (1992).
Hutchings, M.I., Hong, H.J. & Buttner, M.J. Mol. Microbiol. 59, 923–935 (2006).
Hong, H.J., Hutchings, M.I. & Buttner, M.J. Adv. Exp. Med. Biol. 631, 200–213 (2008).
Handwerger, S. & Kolokathis, A. FEMS Microbiol. Lett. 58, 167–170 (1990).
Allen, N.E. & Hobbs, J.N. Jr. FEMS Microbiol. Lett. 132, 107–114 (1995).
Baptista, M., Depardieu, F., Courvalin, P. & Arthur, M. Antimicrob. Agents Chemother. 40, 2291–2295 (1996).
Lai, M.H. & Kirsch, D.R. Antimicrob. Agents Chemother. 40, 1645–1648 (1996).
Ulijasz, A.T., Grenader, A. & Weisblum, B. J. Bacteriol. 178, 6305–6309 (1996).
Grissom-Arnold, J., Alborn, W.E. Jr., Nicas, T.I. & Jaskunas, S.R. Microb. Drug Resist. 3, 53–64 (1997).
Mani, N. et al. J. Antibiot. (Tokyo) 51, 471–479 (1998).
Arthur, M., Depardieu, F. & Courvalin, P. Microbiology 145, 1849–1858 (1999).
Prestwich, G.D., Dorman, G., Elliott, J.T., Marecak, D.M. & Chaudhary, A. Photochem. Photobiol. 65, 222–234 (1997).
Van Bambeke, F., Van Laethem, Y., Courvalin, P. & Tulkens, P.M. Drugs 64, 913–936 (2004).
Neu, J.M. & Wright, G.D. FEMS Microbiol. Lett. 199, 15–20 (2001).
Schwalbe, R.S. et al. Antimicrob. Agents Chemother. 40, 2416–2419 (1996).
Sinha Roy, R. et al. Chem. Biol. 8, 1095–1106 (2001).
Acknowledgements
We thank R. Epand for technical assistance with isothermal titration calorimetry and G. Saalbach (John Innes Centre Proteomics Facility) for the Orbitrap analysis. D. Kahne and C. Walsh provided helpful analysis of preliminary data. This research is funded in part by a Canadian Institutes of Health Research grant (MT-14981), a Canada Research Chair in Biochemistry (to G.D.W.), Biotechnology and Biological Sciences Research Council of the UK (BBSRC) grant 208/P20040 (to H.-J.H. and M.J.B.) and a grant-in-aid to the John Innes Centre from the BBSRC. H.-J.H. acknowledges support from the Royal Society and the Medical Research Council (UK).
Author information
Authors and Affiliations
Contributions
K.K. synthesized the compounds, performed labeling experiments and prepared the figures. H.-J.H. prepared genetic constructs and performed the gene expression studies. X.D.W. prepared genetic constructs. I.N. prepared genetic constructs and assisted in protein preparation. M.J.B. and G.D.W. designed and analyzed the experiments. D.H. performed the NMR. M.J.N. performed the mass spectrometry. G.D.W. wrote the manuscript, and all authors edited it.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Results (PDF 1762 kb)
Rights and permissions
About this article
Cite this article
Koteva, K., Hong, HJ., Wang, X. et al. A vancomycin photoprobe identifies the histidine kinase VanSsc as a vancomycin receptor. Nat Chem Biol 6, 327–329 (2010). https://doi.org/10.1038/nchembio.350
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.350
This article is cited by
-
An overview on the two-component systems of Streptomyces coelicolor
World Journal of Microbiology and Biotechnology (2023)
-
The Extracellular Domain of Two-component System Sensor Kinase VanS from Streptomyces coelicolor Binds Vancomycin at a Newly Identified Binding Site
Scientific Reports (2020)
-
Genetic insights into the mechanism of teicoplanin self-resistance in Actinoplanes teichomyceticus
The Journal of Antibiotics (2020)
-
Histidine kinases and the missing phosphoproteome from prokaryotes to eukaryotes
Laboratory Investigation (2018)
-
Multidrug efflux pumps: structure, function and regulation
Nature Reviews Microbiology (2018)