Abstract
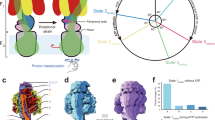
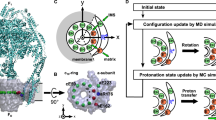
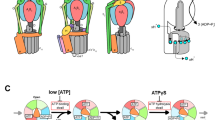
The rotary motor enzyme F1-ATPase (F1) is a catalytic subcomplex of FoF1-ATP synthase that produces most of the ATP in respiring cells. Chemomechanical coupling has been studied extensively for bacterial F1 but very little for mitochondrial F1. Here we report ATP-driven rotation of human mitochondrial F1. A rotor-shaft γ-subunit in the stator α3β3 ring rotates 120° per ATP with three catalytic steps: ATP binding to one β-subunit at 0°, inorganic phosphate (Pi) release from another β-subunit at 65° and ATP hydrolysis on the third β-subunit at 90°. Rotation is often interrupted at 90° by persistent ADP binding and is stalled at 65° by a specific inhibitor azide. A mitochondrial endogenous inhibitor for FoF1-ATP synthase, IF1, blocks rotation at 90°. These features differ from those of bacterial F1, in which both ATP hydrolysis and Pi release occur at around 80°, demonstrating that chemomechanical coupling angles of the γ-subunit are tuned during evolution.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Yoshida, M., Muneyuki, E. & Hisabori, T. ATP synthase—a marvellous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2, 669–677 (2001).
Boyer, P.D. A research journey with ATP synthase. J. Biol. Chem. 277, 39045–39061 (2002).
Senior, A.E., Nadanaciva, S. & Weber, J. The molecular mechanism of ATP synthesis by F1F0-ATP synthase. Biochim. Biophys. Acta 1553, 188–211 (2002).
Noji, H., Yasuda, R., Yoshida, M. & Kinosita, K. Jr. Direct observation of the rotation of F1-ATPase. Nature 386, 299–302 (1997).
Yasuda, R., Noji, H., Kinosita, K. Jr. & Yoshida, M. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120 degree steps. Cell 93, 1117–1124 (1998).
Yasuda, R., Noji, H., Yoshida, M., Kinosita, K. Jr. & Itoh, H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410, 898–904 (2001).
Adachi, K. et al. Coupling of rotation and catalysis in F1-ATPase revealed by single-molecule imaging and manipulation. Cell 130, 309–321 (2007).
Furuike, S. et al. Axle-less F1-ATPase rotates in the correct direction. Science 319, 955–958 (2008).
Watanabe, R. & Noji, H. Timing of inorganic phosphate release modulates the catalytic activity of ATP-driven rotary motor protein. Nat. Commun. 5, 3486 (2014).
Feniouk, B.A., Kato-Yamada, Y., Yoshida, M. & Suzuki, T. Conformational transitions of subunit epsilon in ATP synthase from thermophilic Bacillus PS3. Biophys. J. 98, 434–442 (2010).
Saita, E. et al. Activation and stiffness of the inhibited states of F1-ATPase probed by single-molecule manipulation. J. Biol. Chem. 285, 11411–11417 (2010).
Noji, H. et al. Rotation of Escherichia coli F1-ATPase. Biochem. Biophys. Res. Commun. 260, 597–599 (1999).
Omote, H. et al. The γ-subunit rotation and torque generation in F1-ATPase from wild-type or uncoupled mutant Escherichia coli. Proc. Natl. Acad. Sci. USA 96, 7780–7784 (1999).
Spetzler, D. et al. Single molecule measurements of F1-ATPase reveal an interdependence between the power stroke and the dwell duration. Biochemistry 48, 7979–7985 (2009).
Bilyard, T. et al. High-resolution single-molecule characterization of the enzymatic states in Escherichia coli F1-ATPase. Phil. Trans. R. Soc. Lond. B 368, 20120023 (2013).
Hong, S. & Pedersen, P.L. ATP synthase and the actions of inhibitors utilized to study its roles in human health, disease, and other scientific areas. Microbiol. Mol. Biol. Rev. 72, 590–641 (2008).
Cabezon, E., Butler, P.J., Runswick, M.J. & Walker, J.E. Modulation of the oligomerization state of the bovine F1-ATPase inhibitor protein, IF1, by pH. J. Biol. Chem. 275, 25460–25464 (2000).
Campanella, M. et al. Regulation of mitochondrial structure and function by the F1Fo-ATPase inhibitor protein, IF1. Cell Metab. 8, 13–25 (2008).
Fujikawa, M., Imamura, H., Nakamura, J. & Yoshida, M. Assessing actual contribution of IF1, inhibitor of mitochondrial FoF1, to ATP homeostasis, cell growth, mitochondrial morphology, and cell viability. J. Biol. Chem. 287, 18781–18787 (2012).
Magariyama, Y. et al. Very fast flagellar rotation. Nature 371, 752 (1994).
Shimabukuro, K. et al. Catalysis and rotation of F1 motor: cleavage of ATP at the catalytic site occurs in 1 ms before 40 degree substep rotation. Proc. Natl. Acad. Sci. USA 100, 14731–14736 (2003).
Hirono-Hara, Y. et al. Pause and rotation of F1-ATPase during catalysis. Proc. Natl. Acad. Sci. USA 98, 13649–13654 (2001).
Hirono-Hara, Y., Ishizuka, K., Kinosita, K. Jr., Yoshida, M. & Noji, H. Activation of pausing F1 motor by external force. Proc. Natl. Acad. Sci. USA 102, 4288–4293 (2005).
Sielaff, H. et al. Domain compliance and elastic power transmission in rotary FoF1-ATPase. Proc. Natl. Acad. Sci. USA 105, 17760–17765 (2008).
Okuno, D., Iino, R. & Noji, H. Stiffness of γ subunit of F1-ATPase. Eur. Biophys. J. 39, 1589–1596 (2010).
Martin, J.L., Ishmukhametov, R., Hornung, T., Ahmad, Z. & Frasch, W.D. Anatomy of F1-ATPase powered rotation. Proc. Natl. Acad. Sci. USA 111, 3715–3720 (2014).
Vasilyeva, E.A., Minkov, I.B., Fitin, A.F. & Vinogradov, A.D. Kinetic mechanism of mitochondrial adenosine triphosphatase. Inhibition by azide and activation by sulphite. Biochem. J. 202, 15–23 (1982).
Muneyuki, E. et al. Inhibitory effect of NaN3 on the F0F1 ATPase of submitochondrial particles as related to nucleotide binding. Biochim. Biophys. Acta 1144, 62–68 (1993).
Hyndman, D.J., Milgrom, Y.M., Bramhall, E.A. & Cross, R.L. Nucleotide-binding sites on Escherichia coli F1-ATPase. Specificity of noncatalytic sites and inhibition at catalytic sites by MgADP. J. Biol. Chem. 269, 28871–28877 (1994).
Okuno, D. et al. Correlation between the conformational states of F1-ATPase as determined from its crystal structure and single-molecule rotation. Proc. Natl. Acad. Sci. USA 105, 20722–20727 (2008).
Kabaleeswaran, V., Puri, N., Walker, J.E., Leslie, A.G. & Mueller, D.M. Novel features of the rotary catalytic mechanism revealed in the structure of yeast F1 ATPase. EMBO J. 25, 5433–5442 (2006).
Braig, K., Menz, R.I., Montgomery, M.G., Leslie, A.G. & Walker, J.E. Structure of bovine mitochondrial F1-ATPase inhibited by Mg2+ ADP and aluminium fluoride. Structure 8, 567–573 (2000).
Kagawa, R., Montgomery, M.G., Braig, K., Leslie, A.G. & Walker, J.E. The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride. EMBO J. 23, 2734–2744 (2004).
Lutter, R. et al. Crystallization of F1-ATPase from bovine heart mitochondria. J. Mol. Biol. 229, 787–790 (1993).
Abrahams, J.P., Leslie, A.G., Lutter, R. & Walker, J.E. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 (1994).
Bowler, M.W., Montgomery, M.G., Leslie, A.G. & Walker, J.E. How azide inhibits ATP hydrolysis by the F-ATPases. Proc. Natl. Acad. Sci. USA 103, 8646–8649 (2006).
Abrahams, J.P. et al. The structure of bovine F1-ATPase complexed with the peptide antibiotic efrapeptin. Proc. Natl. Acad. Sci. USA 93, 9420–9424 (1996).
van Raaij, M.J., Abrahams, J.P., Leslie, A.G. & Walker, J.E. The structure of bovine F1-ATPase complexed with the antibiotic inhibitor aurovertin B. Proc. Natl. Acad. Sci. USA 93, 6913–6917 (1996).
Gledhill, J.R., Montgomery, M.G., Leslie, A.G. & Walker, J.E. Mechanism of inhibition of bovine F1-ATPase by resveratrol and related polyphenols. Proc. Natl. Acad. Sci. USA 104, 13632–13637 (2007).
Cingolani, G. & Duncan, T.M. Structure of the ATP synthase catalytic complex (F1) from Escherichia coli in an autoinhibited conformation. Nat. Struct. Mol. Biol. 18, 701–707 (2011).
Bowler, M.W., Montgomery, M.G., Leslie, A.G. & Walker, J.E. Ground state structure of F1-ATPase from bovine heart mitochondria at 1.9 Å resolution. J. Biol. Chem. 282, 14238–14242 (2007).
Knowles, A.F. & Penefsky, H.S. The subunit structure of beef heart mitochondrial adenosine triphosphatase. Isolation procedures. J. Biol. Chem. 247, 6617–6623 (1972).
Suzuki, T., Ueno, H., Mitome, N., Suzuki, J. & Yoshida, M. Fo of ATP synthase is a rotary proton channel. Obligatory coupling of proton translocation with rotation of c-subunit ring. J. Biol. Chem. 277, 13281–13285 (2002).
Gupta, S., Grasnoff, S.B., Roberts, D.W. & Renwick, J.A.A. Structure of efrapeptins from the fungus Tolypocladium niveum: peptide inhibitors of mitochondrial ATPase. J. Org. Chem. 57, 2306–2313 (1992).
Walker, J.E. et al. Primary structure and subunit stoichiometry of F1-ATPase from bovine mitochondria. J. Mol. Biol. 184, 677–701 (1985).
Furuike, S. et al. Resolving stepping rotation in Thermus thermophilus H+-ATPase/synthase with an essentially drag-free probe. Nat. Commun. 2, 233 (2011).
Suzuki, T., Suzuki, J., Mitome, N., Ueno, H. & Yoshida, M. Second stalk of ATP synthase. Cross-linking of γ subunit in F1 to truncated Fob subunit prevents ATP hydrolysis. J. Biol. Chem. 275, 37902–37906 (2000).
Acknowledgements
We thank our colleagues B. Feniouk, N. Taniguchi and K. Sugawara (Japan Science and Technology Agency), S. Furuike (Osaka Medical college), K. Kinosita Jr., N. Soga and K. Adachi (Waseda University) and T. Hisabori (Tokyo Institute of Technology) for their valuable discussions. S. Furuike helped T.S. in setting up the center-shielded dark-field microscopic system. We thank K. Adachi (Waseda University) for the kind gift of the software program Center-5. The cDNA library from human lung cells was a kind gift from F. Ishikawa of Kyoto University and M. Fujikawa of the Japan Science and Technology Agency. This work was supported in part by grant-in-aid for Scientific Research from the Japan Society for the Promotion of Science (grant numbers 24570149 (T.S.) and 23227006 (M.Y.)).
Author information
Authors and Affiliations
Contributions
T.S. and M.Y. conceived and designed the experiments and wrote the paper. T.S. led development of the human F1 expression system in E. coli and the single-molecule observation system. E.S. gave critical suggestions for experiments and interpretations. K.T. and C.W. helped T.S. in collecting large amounts of image data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results and Supplementary Figures 1–7. (PDF 3445 kb)
Supplementary Video 1
Stepwise rotation of human F1. A sample of rotations of human F1 under conditions in Figure 2a (4 mM ATPγS, 50 kfps). (AVI 3004 kb)
Supplementary Video 2
Stepwise rotation of human F1. A sample of rotations of human F1 under conditions in Figure 2b (50 μM ATPγS, 50 kfps). (AVI 3004 kb)
Rights and permissions
About this article
Cite this article
Suzuki, T., Tanaka, K., Wakabayashi, C. et al. Chemomechanical coupling of human mitochondrial F1-ATPase motor. Nat Chem Biol 10, 930–936 (2014). https://doi.org/10.1038/nchembio.1635
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1635
This article is cited by
-
Molecular mechanism on forcible ejection of ATPase inhibitory factor 1 from mitochondrial ATP synthase
Nature Communications (2023)
-
Scattering imaging of biomolecules with metallic nanoparticles: localization precision, imaging speed, and multicolor imaging capability
Optical Review (2022)
-
The catalytic dwell in ATPases is not crucial for movement against applied torque
Nature Chemistry (2020)
-
Correlation between the numbers of rotation steps in the ATPase and proton-conducting domains of F- and V-ATPases
Biophysical Reviews (2020)
-
Efficiencies of molecular motors: a comprehensible overview
Biophysical Reviews (2020)