Abstract
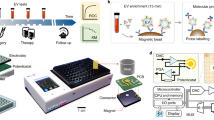
The analysis of cell-free nucleic acids (cfNAs), which are present at significant levels in the blood of cancer patients, can reveal the mutational spectrum of a tumour without the need for invasive sampling of the tissue. However, this requires differentiation between the nucleic acids that originate from healthy cells and the mutated sequences shed by tumour cells. Here we report an electrochemical clamp assay that directly detects mutated sequences in patient serum. This is the first successful detection of cfNAs without the need for enzymatic amplification, a step that normally requires extensive sample processing and is prone to interference. The new chip-based assay reads out the presence of mutations within 15 minutes using a collection of oligonucleotides that sequester closely related sequences in solution, and thus allow only the mutated sequence to bind to a chip-based sensor. We demonstrate excellent levels of sensitivity and specificity and show that the clamp assay accurately detects mutated sequences in a collection of samples taken from lung cancer and melanoma patients.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Stroun, M., Anker, P., Lyautey, J., Lederrey, C. & Maurice, P. A. Isolation and characterization of DNA from the plasma of cancer-patients. Eur. J. Cancer Clin. Oncol. 23, 707–712 (1987).
Kaiser, J. Keeping tabs on tumor DNA. Science 327, 1074 (2010).
Newman, A. M. et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nature Med. 20, 548–556 (2014).
Schwarzenbach, H., Hoon, D. S. B. & Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nature Rev. Cancer 11, 426–437 (2011).
Thierry, A. R. et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nature Med. 20, 430–436 (2014).
Huber, F., Lang, H. P., Backmann, N., Rimoldi, D. & Gerber, C. Direct detection of a BRAF mutation in total RNA from melanoma cells using cantilever arrays. Nature Nanotechnol. 8, 125–129 (2013).
Diehl, F. et al. Circulating mutant DNA to assess tumor dynamics. Nature Med. 14, 985–990 (2008).
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra24 (2014).
Murtaza, M. et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature 497, 108–113 (2013).
Ørum, H. et al. Single base pair mutation analysis by PNA directed PCR clamping. Nucleic Acids Res. 21, 5332–5336 (1993).
Taback, A. et al. Peptide nucleic acid clamp PCR: a novel K-ras mutation detection assay for colorectal cancer micrometastases in lymph nodes. Int. J. Cancer 111, 409–414 (2004).
Chiou, C-C., Luo, J-D. & Chen, T-L. Single-tube reaction using peptide nucleic acid as both PCR clamp and sensor probe for the detection of rare mutations. Nature Protocols 1, 2604–2612 (2007).
García-Olmo, D. C. et al. Colorectal cancer patients induce the oncogenic transformation of susceptible cultured cells. Cancer Res. 70, 560–567 (2010).
Kelley, S. O. et al. Advancing the speed, sensitivity and accuracy of biomolecular detection using multi-length-scale engineering. Nature Nanotechnol. 9, 969–980 (2014).
Bakker, E. & Qin, Y. Electrochemical sensors. Anal. Chem. 78, 3965–3983 (2006).
Das, J. & Kelley, S. O. Protein detection using arrayed microsensor chips: tuning sensor footprint to achieve ultrasensitive readout of CA-125 in serum and whole blood. Anal. Chem. 83, 1167–1172 (2011).
Wen, Y. et al. DNA nanostructure-based interfacial engineering for PCR-free ultrasensitive electrochemical analysis of microRNA. Sci. Rep. 2, 867 (2012).
Chuah, K. et al. Ultrasensitive electrochemical detection of prostate-specific antigen (PSA) using gold-coated magnetic nanoparticles as ‘dispersible electrodes’. Chem. Commun. 48, 3503–3505 (2012).
Si, Y. et al. Ultrasensitive electroanalysis of low-level free microRNAs in blood by maximum signal amplification of catalytic silver deposition using alkaline phosphatase-incorporated gold nanoclusters. Anal. Chem. 86, 10406–10414 (2014).
Rusling, J. F. Multiplexed electrochemical protein detection and translation to personalized cancer diagnostics. Anal. Chem. 85, 5304–5310 (2013).
Fang, Z. et al. Direct profiling of cancer biomarkers in tumour tissue using a multiplexed nanostructured microelectrode integrated circuit. ACS Nano 3, 3207–3213 (2009).
Soleymani, L. et al. Hierarchical nanotextured microelectrodes overcome the molecular transport barrier to achieve rapid, direct bacterial detection. ACS Nano 5, 3360–3366 (2011).
Ferguson, B. S. et al. Genetic analysis of H1N1 influenza virus from throat swab samples in a microfluidic system for point-of-care diagnostics. J. Am. Chem. Soc. 133, 9129–9135 (2011).
Hsieh, K., Patterson, A. S., Ferguson, B. S., Plaxco, K. W. & Soh, H. T. Rapid, sensitive, and quantitative detection of pathogenic DNA at the point of care through microfluidic electrochemical quantitative loop-mediated isothermal amplification. Angew. Chem. Int. Ed. 51, 4896–4900 (2012).
Yang, H. et al. Direct, electronic microRNA detection for the rapid determination of differential expression profiles. Angew. Chem. Int. Ed. 48, 8461–8464 (2009).
Wu, Y. & Lai, R. Y. Development of a ‘signal-on’ electrochemical DNA sensor with an oligo-thymine spacer for point mutation detection. Chem. Commun. 49, 3422–3424 (2013).
Wee, E. J. H., Shiddiky, M. J. A., Brown, M. A. & Trau, M. ELCR: electrochemical detection of single DNA base changes via ligase chain reaction. Chem. Commun. 48, 12014–12016 (2012).
Xiang, Y. & Lu, Y. Using personal glucose meters and functional DNA sensors to quantify a variety of analytical targets. Nature Chem. 3, 697–703 (2011).
Drummond, T. G., Hill, M. G. & Barton, J. K. Electrochemical DNA sensors. Nature Biotechnol. 21, 1192–1199 (2003).
Ge, Z. et al. Hybridization chain reaction amplification of microRNA detection with a tetrahedral DNA nanostructure-based electrochemical biosensor. Anal. Chem. 86, 2124–2130 (2014).
Hsieh, K., Patterson, A. S., Ferguson, B. S., Plaxco, K. W. & Soh, H. T. Rapid, sensitive, and quantitative detection of pathogenic DNA at the point of care via microfluidic electrochemical quantitative loop-mediated isothermal amplification (MEQ-LAMP). Angew. Chem. Int. Ed. 124, 4980–4984 (2012).
Tavallaie, R., Darwish, N., Gebala, M., Hibbert, D. B. & Gooding, J. J. The effect of interfacial design on the electrochemical detection of DNA and microRNA using methylene blue at low-density DNA films. ChemElectroChem 1, 165–171 (2014).
Gautschi, O. et al. Origin and prognostic value of circulating KRAS mutations in lung cancer patients. Cancer Lett. 254, 265–273 (2007).
Wang, S. et al. Potential clinical significance of a plasma-based KRAS mutation analysis in patients with advanced non-small cell lung cancer. Clin. Cancer Res. 16, 1324–1330 (2010).
Soleymani, L., Fang, Z., Sargent, E. H. & Kelley, S. O. Programming the detection limits of biosensors through controlled nanostructuring. Nature Nanotechnol. 4, 844–848 (2009).
Soleymani, L. et al. Nanostructuring of patterned microelectrodes to enhance the sensitivity of electrochemical nucleic acids detection. Angew. Chem. Int. Ed. 48, 8457–8460 (2009).
Das, J. et al. An ultrasensitive universal detector based on neutralizer displacement. Nature Chem. 4, 642–648 (2012).
Das, J. & Kelley, S. O. Tuning the bacterial detection sensitivity of nanostructured microelectrodes. Anal. Chem. 85, 7333–7338 (2013).
Lam, B. et al. Solution-based circuits enable rapid and multiplexed pathogen detection. Nature Commun. 4, 2001 (2013).
Besant, J. D., Das, J., Sargent, E. H. & Kelley, S. O. Proximal bacterial lysis and detection in nanoliter wells using electrochemistry. ACS Nano 7, 8183–8189 (2013).
Bin, X., Sargent, E. H. & Kelley, S. O. Nanostructuring of sensors determines the efficiency of biomolecular capture. Anal. Chem. 82, 5928–5931 (2010).
Lapierre, M. A., O'Keefe, M. M., Taft, B. J. & Kelley, S. O. Electrocatalytic detection of pathogenic DNA sequences and antibiotic resistance markers. Anal. Chem. 75, 6327–6333 (2003).
PNAClamp KRAS Mutation Detection Kit (Ver.2), Instruction Manual for Product #PNAC-1002 Version 4.1 (PNAGENE, 2012).
PNAClamp BRAF Mutation Detection Kit, Instruction Manual for Product #PNAC-2001 Version 4.4 (PNAGENE, 2012).
Abe, K. Direct PCR from serum: application to viral genome detection in PCR Protocols (eds Bartlett, J. M. S. & Stirling, D.) 161–166 (Tatowa, 2003).
Zhou, Y., Wan, Y., Sage, A., Poudineh, M. & Kelley, S. O. Effect of microelectrode structure on electrocatalysis at nucleic acid-modified sensors. Langmuir 30, 14322–14328 (2014).
Acknowledgements
This research was sponsored by the Ontario Research Fund (Research Excellence Award to S.O.K.), the Canadian Institutes for Health Research (Emerging Team Grant to S.O.K. and E.H.S.), the Canadian Cancer Society Research Institute (Innovation Grant No. 702414 to S.O.K. and J.R.) and the Natural Science and Engineering Research Council (Discovery Grant to S.O.K.).
Author information
Authors and Affiliations
Contributions
J.D., I.I., J.R., E.H.S. and S.O.K. conceived the experiments, J.D. and I.I. designed the experiments, L.M. contributed critical materials and J.D., I.I., E.H.S. and S.O.K. co-wrote the paper. All the authors reviewed and improved the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 1293 kb)
Rights and permissions
About this article
Cite this article
Das, J., Ivanov, I., Montermini, L. et al. An electrochemical clamp assay for direct, rapid analysis of circulating nucleic acids in serum. Nature Chem 7, 569–575 (2015). https://doi.org/10.1038/nchem.2270
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.2270
This article is cited by
-
Electrochemical biosensors for analysis of DNA point mutations in cancer research
Analytical and Bioanalytical Chemistry (2023)
-
Limitations and opportunities of technologies for the analysis of cell-free DNA in cancer diagnostics
Nature Biomedical Engineering (2022)
-
An integrated magneto-electrochemical device for the rapid profiling of tumour extracellular vesicles from blood plasma
Nature Biomedical Engineering (2021)
-
A deep learning model for predicting next-generation sequencing depth from DNA sequence
Nature Communications (2021)
-
Long non-coding RNA CCAT2 as a potential serum biomarker for diagnosis and prognosis of multiple myeloma
Annals of Hematology (2020)