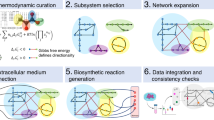
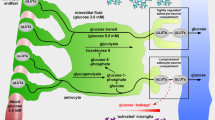
Abstract
Metabolic interactions between multiple cell types are difficult to model using existing approaches. Here we present a workflow that integrates gene expression data, proteomics data and literature-based manual curation to model human metabolism within and between different types of cells. Transport reactions are used to account for the transfer of metabolites between models of different cell types via the interstitial fluid. We apply the method to create models of brain energy metabolism that recapitulate metabolic interactions between astrocytes and various neuron types relevant to Alzheimer's disease. Analysis of the models identifies genes and pathways that may explain observed experimental phenomena, including the differential effects of the disease on cell types and regions of the brain. Constraint-based modeling can thus contribute to the study and analysis of multicellular metabolic processes in the human tissue microenvironment and provide detailed mechanistic insight into high-throughput data analysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Feist, A.M. & Palsson, B.Ø. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat. Biotechnol. 26, 659–667 (2008).
Oberhardt, M.A., Palsson, B.Ø. & Papin, J.A. Applications of genome-scale metabolic reconstructions. Mol. Syst. Biol. 5, 320 (2009).
Breitling, R., Vitkup, D. & Barrett, M.P. New surveyor tools for charting microbial metabolic maps. Nat. Rev. Microbiol. 6, 156–161 (2008).
Thiele, I. & Palsson, B.O. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 5, 93–121 (2010).
Lewis, N.E., Jamshidi, N., Thiele, I. & Palsson, B.Ø. in Encyclopedia of Complexity and Systems Science (ed. Meyers, R.A.) 5535–5552 (Springer, New York, 2009).
Orth, J.D., Thiele, I. & Palsson, B.O. What is flux balance analysis? Nat. Biotechnol. 28, 245–248 (2010).
Duarte, N.C. et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc. Natl. Acad. Sci. USA 104, 1777–1782 (2007).
Becker, S.A. & Palsson, B.O. Context-specific metabolic networks are consistent with experiments. PLOS Comput. Biol. 4, e1000082 (2008).
Shlomi, T., Cabili, M.N., Herrgard, M.J., Palsson, B.Ø. & Ruppin, E. Network-based prediction of human tissue–specific metabolism. Nat. Biotechnol. 26, 1003–1010 (2008).
Jerby, L., Shlomi, T. & Ruppin, E. Computational reconstruction of tissue-specific metabolic models: application to human liver metabolism. Mol. Syst. Biol. 6, 401 (2010).
Ponten, F. et al. A global view of protein expression in human cells, tissues, and organs. Mol. Syst. Biol. 5, 337 (2009).
Palsson, B.O. in Systems Biology: Properties of Reconstructed Networks 322 (Cambridge University Press, Cambridge/New York, 2006).
Mishra, G.R. et al. Human protein reference database–2006 update. Nucleic Acids Res. 34, D411–D414 (2006).
Fujii, Y., Imanishi, T. & Gojobori, T. H-Invitational Database: integrated database of human genes. Tanpakushitsu Kakusan Koso 49, 1937–1943 (2004).
Reidegeld, K.A. et al. The power of cooperative investigation: summary and comparison of the HUPO Brain Proteome Project pilot study results. Proteomics 6, 4997–5014 (2006).
Chatziioannou, A., Palaiologos, G. & Kolisis, F.N. Metabolic flux analysis as a tool for the elucidation of the metabolism of neurotransmitter glutamate. Metab. Eng. 5, 201–210 (2003).
Cakir, T., Alsan, S., Saybasili, H., Akin, A. & Ulgen, K.O. Reconstruction and flux analysis of coupling between metabolic pathways of astrocytes and neurons: application to cerebral hypoxia. Theor. Biol. Med. Model. 4, 48 (2007).
Occhipinti, R., Puchowicz, M.A., LaManna, J.C., Somersalo, E. & Calvetti, D. Statistical analysis of metabolic pathways of brain metabolism at steady state. Ann. Biomed. Eng. 35, 886–902 (2007).
Reiman, E.M. et al. Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer's dementia. Proc. Natl. Acad. Sci. USA 101, 284–289 (2004).
Ginsberg, S.D., Che, S., Counts, S.E. & Mufson, E.J. Single cell gene expression profiling in Alzheimer's disease. NeuroRx 3, 302–318 (2006).
Lai, M.K.P., Ramirez, M.J., Tsang, S.W.Y. & Francis, P.T. Alzheimer's disease as a neurotransmitter disease in Neurobiology of Alzheimer' Disease (eds. Dawbarn, D. & Allen, S.J.) 245–281 (Oxford University Press, New York, 2007).
Fukui, H., Diaz, F., Garcia, S. & Moraes, C.T. Cytochrome c oxidase deficiency in neurons decreases both oxidative stress and amyloid formation in a mouse model of Alzheimer's disease. Proc. Natl. Acad. Sci. USA 104, 14163–14168 (2007).
Bubber, P., Haroutunian, V., Fisch, G., Blass, J.P. & Gibson, G.E. Mitochondrial abnormalities in Alzheimer brain: mechanistic implications. Ann. Neurol. 57, 695–703 (2005).
Gibson, G.E. et al. Alpha-ketoglutarate dehydrogenase in Alzheimer brains bearing the APP670/671 mutation. Ann. Neurol. 44, 676–681 (1998).
Casley, C.S., Canevari, L., Land, J.M., Clark, J.B. & Sharpe, M.A. Beta-amyloid inhibits integrated mitochondrial respiration and key enzyme activities. J. Neurochem. 80, 91–100 (2002).
Hoshi, M. et al. Regulation of mitochondrial pyruvate dehydrogenase activity by tau protein kinase I/glycogen synthase kinase 3beta in brain. Proc. Natl. Acad. Sci. USA 93, 2719–2723 (1996).
Gorman, A.M., Ceccatelli, S. & Orrenius, S. Role of mitochondria in neuronal apoptosis. Dev. Neurosci. 22, 348–358 (2000).
Santos, S.S. et al. Inhibitors of the alpha-ketoglutarate dehydrogenase complex alter [1–13C]glucose and [U-13C]glutamate metabolism in cerebellar granule neurons. J. Neurosci. Res. 83, 450–458 (2006).
Hassel, B., Johannessen, C.U., Sonnewald, U. & Fonnum, F. Quantification of the GABA shunt and the importance of the GABA shunt versus the 2-oxoglutarate dehydrogenase pathway in GABAergic neurons. J. Neurochem. 71, 1511–1518 (1998).
Liang, W.S. et al. Altered neuronal gene expression in brain regions differentially affected by Alzheimer's disease: a reference data set. Physiol. Genomics 33, 240–256 (2008).
Stuhmer, T., Anderson, S.A., Ekker, M. & Rubenstein, J.L. Ectopic expression of the Dlx genes induces glutamic acid decarboxylase and Dlx expression. Development 129, 245–252 (2002).
Ibanez, V. et al. Regional glucose metabolic abnormalities are not the result of atrophy in Alzheimer's disease. Neurology 50, 1585–1593 (1998).
Schramm, G. et al. PathWave: discovering patterns of differentially regulated enzymes in metabolic pathways. Bioinformatics 26, 1225–1231 (2010).
Ragozzino, M.E., Pal, S.N., Unick, K., Stefani, M.R. & Gold, P.E. Modulation of hippocampal acetylcholine release and spontaneous alternation scores by intrahippocampal glucose injections. J. Neurosci. 18, 1595–1601 (1998).
Watson, G.S. & Craft, S. Modulation of memory by insulin and glucose: neuropsychological observations in Alzheimer's disease. Eur. J. Pharmacol. 490, 97–113 (2004).
Cooper, J.R. Unsolved problems in the cholinergic nervous system. J. Neurochem. 63, 395–399 (1994).
Karczmar, A.G. Cholinergic cells and pathways in Exploring the Vertebrate Cholinergic Nervous System 686 (Springer, New York, 2006).
Gibson, G.E., Jope, R. & Blass, J.P. Decreased synthesis of acetylcholine accompanying impaired oxidation of pyruvic acid in rat brain minces. Biochem. J. 148, 17–23 (1975).
Abbott, N.J., Ronnback, L. & Hansson, E. Astrocyte-endothelial interactions at the blood-brain barrier. Nat. Rev. Neurosci. 7, 41–53 (2006).
Thompson, M.D., Knee, K. & Golden, C.J. Olfaction in persons with Alzheimer's disease. Neuropsychol. Rev. 8, 11–23 (1998).
Schryer, D.W., Peterson, P., Paalme, T. & Vendelin, M. Bidirectionality and compartmentation of metabolic fluxes are revealed in the dynamics of isotopomer networks. Int. J. Mol. Sci. 10, 1697–1718 (2009).
Serres, S., Raffard, G., Franconi, J.M. & Merle, M. Close coupling between astrocytic and neuronal metabolisms to fulfill anaplerotic and energy needs in the rat brain. J. Cereb. Blood Flow Metab. 28, 712–724 (2008).
Bordbar, A. et al. Insight into human alveolar macrophage and M. tuberculosis interactions via metabolic reconstructions. Mol. Syst. Biol. 6, 422 (2010).
Drug off-target effects predicted using structural analysis in the context of a metabolic network model. PLoS. Comput. Biol. 6, e1000938 (2010).
Lee, D.S. et al. The implications of human metabolic network topology for disease comorbidity. Proc. Natl. Acad. Sci. USA 105, 9880–9885 (2008).
Palsson, B.O. & Zengler, K. The challenges of integrating multi-omics data sets. Nat. Chem. Biol. 6, 787–789 (2010).
Lying-Tunell, U., Lindblad, B.S., Malmlund, H.O. & Persson, B. Cerebral blood flow and metabolic rate of oxygen, glucose, lactate, pyruvate, ketone bodies and amino acids. Acta Neurol. Scand. 62, 265–275 (1980).
Lying-Tunell, U., Lindblad, B.S., Malmlund, H.O. & Persson, B. Cerebral blood flow and metabolic rate of oxygen, glucose, lactate, pyruvate, ketone bodies and amino acids. Acta Neurol. Scand. 63, 337–350 (1981).
Tischfield, M.A. et al. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell 140, 74–87 (2010).
Kim, K.K., Adelstein, R.S. & Kawamoto, S. Identification of neuronal nuclei (NeuN) as Fox-3, a new member of the Fox-1 gene family of splicing factors. J. Biol. Chem. 284, 31052–31061 (2009).
De Camilli, P., Cameron, R. & Greengard, P. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. I. Its general distribution in synapses of the central and peripheral nervous system demonstrated by immunofluorescence in frozen and plastic sections. J. Cell Biol. 96, 1337–1354 (1983).
Olave, I., Wang, W., Xue, Y., Kuo, A. & Crabtree, G.R. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev. 16, 2509–2517 (2002).
Benjamini, Y. & Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188 (2001).
Acknowledgements
The authors thank G. Gibson at Cornell University, I. Thiele at the University of Iceland and M. Abrams, M. Mo and C. Barrett at UCSD for suggestions pertaining to this work. This work was funded in part by a Fulbright fellowship, a National Science Foundation IGERT Plant Systems Biology training grant (no. DGE-0504645), US National Institutes of Health grants 2R01GM068837_05A1 and RO1 GM071808 and the Helmholtz Alliance on Systems Biology and the BMBF by the NGFN+ neuroblastoma project ENGINE.
Author information
Authors and Affiliations
Contributions
N.E.L., J.K.C., A.Y., N.P., M.P.A. and B.O.P. conceived and designed the model. N.E.L., J.K.C., G.S., R.K., R.E., J.S., A.B. and R.A.L. performed data analyses. The manuscript was written by N.E.L., G.S., J.S., A.B. and B.O.P.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Notes (PDF 1971 kb)
Supplementary Model 1
All neuron types normal (ZIP 72 kb)
Supplementary Model 2
All neuron types normal elderly (ZIP 72 kb)
Supplementary Model 3
All neuron types Alzheimer's (ZIP 72 kb)
Supplementary Table 1
Model rxns (XLS 317 kb)
Supplementary Table 2
Model metabolites (XLS 109 kb)
Supplementary Table 3
Model genes (XLS 50 kb)
Supplementary Table 4
Model S. matrix (ZIP 904 kb)
Supplementary Table 5
Model parameters (XLS 24 kb)
Supplementary Table 6
Significant Pathwave pathways (XLS 25 kb)
Supplementary Table 7
Protein data accession numbers (XLS 18 kb)
Supplementary Table 8
Comparison with other models (XLS 18 kb)
Supplementary Table 9
Pathwave pathway classes (XLS 42 kb)
Supplementary Table 10
Pathwave exception metabolites (XLS 15 kb)
Supplementary Table 11
Significant Pathwave features (XLS 168 kb)
Rights and permissions
About this article
Cite this article
Lewis, N., Schramm, G., Bordbar, A. et al. Large-scale in silico modeling of metabolic interactions between cell types in the human brain. Nat Biotechnol 28, 1279–1285 (2010). https://doi.org/10.1038/nbt.1711
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.1711
This article is cited by
-
Extracting functionally accurate context-specific models of Atlantic salmon metabolism
npj Systems Biology and Applications (2023)
-
Linear programming based gene expression model (LPM-GEM) predicts the carbon source for Bacillus subtilis
BMC Bioinformatics (2022)
-
Developing a dynamic equilibrium system in Escherichia coli to improve the production of recombinant proteins
Applied Microbiology and Biotechnology (2022)
-
Mathematical Framework Behind the Reconstruction and Analysis of Genome Scale Metabolic Models
Archives of Computational Methods in Engineering (2019)
-
A linked organ-on-chip model of the human neurovascular unit reveals the metabolic coupling of endothelial and neuronal cells
Nature Biotechnology (2018)