Abstract
The mammalian gut is colonized by numerous microorganisms collectively termed the microbiota, which have a mutually beneficial relationship with their host1,2,3. Normally, the gut microbiota matures during ontogeny to a state of balanced commensalism marked by the absence of adverse inflammation4,5. Subsets of innate lymphoid cells (ILCs) and conventional T cells are considered to have redundant functions in containment and clearance of microbial pathogens6,7, but how these two major lymphoid-cell populations each contribute to shaping the mature commensal microbiome and help to maintain tissue homeostasis has not been determined. Here we identify, using advanced multiplex quantitative imaging methods, an extensive and persistent phosphorylated-STAT3 signature in group 3 ILCs and intestinal epithelial cells that is induced by interleukin (IL)-23 and IL-22 in mice that lack CD4+ T cells. By contrast, in immune-competent mice, phosphorylated-STAT3 activation is induced only transiently by microbial colonization at weaning. This early signature is extinguished as CD4+ T cell immunity develops in response to the expanding commensal burden. Physiologically, the persistent IL-22 production from group 3 ILCs that occurs in the absence of adaptive CD4+ T-cell activity results in impaired host lipid metabolism by decreasing lipid transporter expression in the small bowel. These findings provide new insights into how innate and adaptive lymphocytes operate sequentially and in distinct ways during normal development to establish steady-state commensalism and tissue metabolic homeostasis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Caballero, S. & Pamer, E. G. Microbiota-mediated inflammation and antimicrobial defense in the intestine. Annu. Rev. Immunol. 33, 227–256 (2015)
Hooper, L. V., Littman, D. R. & Macpherson, A. J. Interactions between the microbiota and the immune system. Science 336, 1268–1273 (2012)
Belkaid, Y. & Hand, T. W. Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014)
Renz, H., Brandtzaeg, P. & Hornef, M. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat. Rev. Immunol. 12, 9–23 (2011)
Gensollen, T., Iyer, S. S., Kasper, D. L. & Blumberg, R. S. How colonization by microbiota in early life shapes the immune system. Science 352, 539–544 (2016)
Artis, D. & Spits, H. The biology of innate lymphoid cells. Nature 517, 293–301 (2015)
Bando, J. K. & Colonna, M. Innate lymphoid cell function in the context of adaptive immunity. Nat. Immunol. 17, 783–789 (2016)
Gerner, M. Y., Kastenmuller, W., Ifrim, I., Kabat, J. & Germain, R. N. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity 37, 364–376 (2012)
Gerner, M. Y., Torabi-Parizi, P. & Germain, R. N. Strategically localized dendritic cells promote rapid T cell responses to lymph-borne particulate antigens. Immunity 42, 172–185 (2015)
Liu, Z. et al. Immune homeostasis enforced by co-localized effector and regulatory T cells. Nature 528, 225–230 (2015)
Nguyen, P. M., Putoczki, T. L. & Ernst, M. STAT3-activating cytokines: a therapeutic opportunity for inflammatory bowel disease? J. Interferon Cytokine Res. 35, 340–350 (2015)
Klose, C. S. et al. A T-bet gradient controls the fate and function of CCR6−RORγt+ innate lymphoid cells. Nature 494, 261–265 (2013)
Ivanov, I. I. et al. Induction of intestinal TH17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009)
Sano, T. et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector TH17 responses. Cell 163, 381–393 (2015)
Guo, X. et al. Induction of innate lymphoid cell-derived interleukin-22 by the transcription factor STAT3 mediates protection against intestinal infection. Immunity 40, 25–39 (2014)
Buonocore, S. et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature 464, 1371–1375 (2010)
Tamoutounour, S. et al. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity 39, 925–938 (2013)
Jiang, H. Q., Bos, N. A. & Cebra, J. J. Timing, localization, and persistence of colonization by segmented filamentous bacteria in the neonatal mouse gut depend on immune status of mothers and pups. Infect. Immun. 69, 3611–3617 (2001)
Goto, Y. et al. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal TH17 cell differentiation. Immunity 40, 594–607 (2014)
Yang, Y. et al. Focused specificity of intestinal TH17 cells towards commensal bacterial antigens. Nature 510, 152–156 (2014)
Eberl, G., Di Santo, J. P. & Vivier, E. The brave new world of innate lymphoid cells. Nat. Immunol. 16, 1–5 (2015)
Fang, D. & Zhu, J. Dynamic balance between master transcription factors determines the fates and functions of CD4 T cell and innate lymphoid cell subsets. J. Exp. Med. 214, 1861–1876 (2017)
Korn, L. L. et al. Conventional CD4+ T cells regulate IL-22-producing intestinal innate lymphoid cells. Mucosal Immunol. 7, 1045–1057 (2014)
Littman, D. R. & Rudensky, A. Y. TH17 and regulatory T cells in mediating and restraining inflammation. Cell 140, 845–858 (2010)
Fontenot, J. D., Rasmussen, J. P., Gavin, M. A. & Rudensky, A. Y. A function for interleukin 2 in FOXP3-expressing regulatory T cells. Nat. Immunol. 6, 1142–1151 (2005)
Wang, X. et al. Interleukin-22 alleviates metabolic disorders and restores mucosal immunity in diabetes. Nature 514, 237–241 (2014). 10.1038/nature13564
Wang, Y. et al. The intestinal microbiota regulates body composition through NFIL3 and the circadian clock. Science 357, 912–916 (2017)
Ibiza, S. et al. Glial-cell-derived neuroregulators control type 3 innate lymphoid cells and gut defence. Nature 535, 440–443 (2016)
Cardoso, V. et al. Neuronal regulation of type 2 innate lymphoid cells via neuromedin U. Nature 549, 277–281 (2017)
Klose, C. S. N. et al. The neuropeptide neuromedin U stimulates innate lymphoid cells and type 2 inflammation. Nature 549, 282–286 (2017)
Blanton, L. V., Barratt, M. J., Charbonneau, M. R., Ahmed, T. & Gordon, J. I. Childhood undernutrition, the gut microbiota, and microbiota-directed therapeutics. Science 352, 1533 (2016)
Subramanian, S. et al. Cultivating healthy growth and nutrition through the gut microbiota. Cell 161, 36–48 (2015)
Qiu, J. et al. Group 3 innate lymphoid cells inhibit T-cell-mediated intestinal inflammation through aryl hydrocarbon receptor signaling and regulation of microflora. Immunity 39, 386–399 (2013)
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013)
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014)
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014)
Acknowledgements
We thank Y. Choi and D.M. Kobuley for providing germ free Rag1−/− mice and performing SFB mono-colonization; M. Oukka and S. K. Durum for providing mice; M. Mack, B. Gao and Y. Umesaki for providing anti-CCR2 antibody, IL-22 adenovirus and SFB faecal pellets; C. Eigsti, V. Nair and J. Davis for cell sorting, scanning electron microscopy and microbiota analysis; J. Zhu for discussions; and members of the Laboratory of Systems Biology for their comments during the course of these studies and input during preparation of this manuscript. Y.H. was supported by an NIAID K99 award (1K99AI123350-01A1). This research was supported by the Intramural Research Program of NIAID, NIH.
Author information
Authors and Affiliations
Contributions
K.M. designed and conducted most of the experiments and data analysis and prepared the manuscript; A.P.B., S.T. and Y.H. helped with cell isolation and transfer; L.Z. measured mouse body composition; N.B. performed the analysis of microbiota translocation; A.J.M. performed the RNA-seq and data analysis; M.Y.G. provided helpful suggestions regarding imaging and histo-cytometry; Y.B. provided helpful suggestions, discussed data interpretation and contributed to the manuscript; and R.N.G. designed experiments, interpreted data and helped to write the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
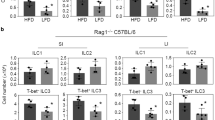
Extended Data Figure 1 Quantification of pSTAT3+ ILC3s by histo-cytometry.
Gating strategy for analysis of pSTAT3+ ILC3s from small intestine of wild-type or Rag1−/− mice.
Extended Data Figure 2 Cellular and molecular mechanism of STAT3 activation in Rag1−/− small intestine.
a, Immunofluorescence staining of ileum from Rag1−/− (n = 4), Il23a−/−Rag1−/− (n = 5), Il22−/−Rag1−/− (n = 4) and Il6−/−Rag1−/− mice (n = 4). b, Percentage of pSTAT3+ ILC3s in a. c–d, Immunofluorescence staining of ileum from Rorc(γt)GFP/+Rag1−/− and Rorc(γt)GFP/GFPRag1−/− mice (n = 4; c) and Il22-tdTomato Rag1−/− mice (n = 3; d). Results are representative of three independent experiments. Bars show mean; exact P values are given and calculated by one-way ANOVA.
Extended Data Figure 3 Mononuclear phagocyte subpopulation responsible for IL-23 production and pSTAT3 activation.
a, Flow cytometry of total live cells from the small intestine lamina propria, showing the gating strategy for sorting different myeloid-cell subsets: CD103+CD11b− and CD11b+ conventional dendritic cells, CD64+CCR2− macrophages and CCR2+ monocytes and monocyte-derived dendritic cells. b, Expression of Il23a (n = 3) and Il12b (which encodes the p40 subunit of IL-12) in different cell populations sorted as in a (n = 2). c, Immunofluorescence staining of ileum from Rag1−/− mice and Rag1−/− mice treated with anti-CCR2 or anti-Gr1 antibody for two weeks (n = 4). Results are representative of three independent experiments. Mean ± s.d.; exact P values are given and calculated by two-way ANOVA.
Extended Data Figure 4 Microorganisms in the small intestine of co-housed wild-type, Rag1−/− and Il23a−/−Rag1−/− mice.
a, Quantification of indicated bacteria species in the ileum of co-housed wild-type (n = 6), Rag1−/− (n = 7) and Il23a−/−Rag1−/− (n = 8) mice by real-time PCR with primers specific to 16S rRNA genes. Results are pooled from two independent experiments. b, Scanning electron microscopy of terminal ileum of co-housed mice as in a (n = 3). c, Quantification of the length of SFB filaments in b. Results are representative of two independent experiments. Bars show mean (a) and mean ± s.d. (c); exact P values are given and calculated by one-way ANOVA.
Extended Data Figure 5 Lack of ILC3 activation in SFB negative Tcra−/− mice.
a, Immunofluorescence staining of ileum from wild-type (n = 6), Rag1−/− (n = 5), Tcra−/− (n = 5) and Ighm−/− mice (n = 6). b, Percentage of pSTAT3+ ILC3s in a. c, Quantification of SFB in distal small intestine of non-co-housed Rag1−/− and Tcra−/− or co-housed Rag1−/− and Tcra−/− mice (n = 4). Results are representative of three (a, b) or two (c) independent experiments. Bars show mean; ****P < 0.0001; otherwise exact P values are shown and calculated by one-way ANOVA.
Extended Data Figure 6 Dysregulation of lipid metabolism by IL-22.
a, Expression of indicated genes in the ileum from age-matched male Tcra−/− mice and Tcra−/− mice co-housed with Rag1−/− mice (n = 8). Results are pooled from two independent experiments. b, Serum triglyceride and free fatty acid levels from Tcra−/− mice and Tcra−/− mice co-housed with Rag1−/− mice (n = 5). c–f, Expression of indicated genes in the ileum (c, e) and serum triglyceride and free fatty acid levels (d, f) from wild-type (c, d) or Tcra−/− mice (e, f) injected with adenovirus expressing IL-22 (n = 5) or GFP (n = 5). Results are representative of two independent experiments. Bars show mean (a, c, e) and mean ± s.d. (b, d, f); exact P values are given and calculated by two-tailed Student’s t-test.
Extended Data Figure 7 STAT3 activation in IECs by IL-22 adenovirus.
Immunofluorescence staining of ileum from C57BL/6 mice injected with adenoviruses expressing either IL-22 or GFP for two weeks. Images are representative of three sections from three mice of each group and results are representative of two independent experiments.
Supplementary information
Rights and permissions
About this article
Cite this article
Mao, K., Baptista, A., Tamoutounour, S. et al. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature 554, 255–259 (2018). https://doi.org/10.1038/nature25437
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25437
This article is cited by
-
Group 3 innate lymphoid cells in intestinal health and disease
Nature Reviews Gastroenterology & Hepatology (2024)
-
Impacts of maternal microbiota and microbial metabolites on fetal intestine, brain, and placenta
BMC Biology (2023)
-
Metabolic control by the microbiome
Genome Medicine (2022)
-
Significant alterations of intestinal symbiotic microbiota induced by intraperitoneal vaccination mediate changes in intestinal metabolism of NEW Genetically Improved Farmed Tilapia (NEW GIFT, Oreochromis niloticus)
Microbiome (2022)
-
Heterogeneity of type 2 innate lymphoid cells
Nature Reviews Immunology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.