Abstract
Protein biosynthesis depends on the availability of ribosomes, which in turn relies on ribosomal RNA production. In eukaryotes, this process is carried out by RNA polymerase I (Pol I), a 14-subunit enzyme, the activity of which is a major determinant of cell growth. Here we present the crystal structure of Pol I from Saccharomyces cerevisiae at 3.0 Å resolution. The Pol I structure shows a compact core with a wide DNA-binding cleft and a tightly anchored stalk. An extended loop mimics the DNA backbone in the cleft and may be involved in regulating Pol I transcription. Subunit A12.2 extends from the A190 jaw to the active site and inserts a transcription elongation factor TFIIS-like zinc ribbon into the nucleotide triphosphate entry pore, providing insight into the role of A12.2 in RNA cleavage and Pol I insensitivity to α-amanitin. The A49–A34.5 heterodimer embraces subunit A135 through extended arms, thereby contacting and potentially regulating subunit A12.2.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Warner, J. R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24, 437–440 (1999)
Drygin, D., Rice, W. G. & Grummt, I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol. 50, 131–156 (2010)
Grummt, I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 17, 1691–1702 (2003)
Moss, T., Langlois, F., Gagnon-Kugler, T. & Stefanovsky, V. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell. Mol. Life Sci. 64, 29–49 (2007)
Vannini, A. & Cramer, P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol. Cell 45, 439–446 (2012)
Russell, J. & Zomerdijk, J. C. The RNA polymerase I transcription machinery. Biochem. Soc. Symp. 73, 203–216 (2006)
Kuhn, C. D. et al. Functional architecture of RNA polymerase I. Cell 131, 1260–1272 (2007)
Ruan, W., Lehmann, E., Thomm, M., Kostrewa, D. & Cramer, P. Evolution of two modes of intrinsic RNA polymerase transcript cleavage. J. Biol. Chem. 286, 18701–18707 (2011)
Carter, R. & Drouin, G. The increase in the number of subunits in eukaryotic RNA polymerase III relative to RNA polymerase II is due to the permanent recruitment of general transcription factors. Mol. Biol. Evol. 27, 1035–1043 (2010)
Geiger, S. R. et al. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol. Cell 39, 583–594 (2010)
Cramer, P. et al. Structure of eukaryotic RNA polymerases. Ann. Rev. Biophys. 37, 337–352 (2008)
Fernández-Tornero, C. et al. Conformational flexibility of RNA polymerase III during transcriptional elongation. EMBO J. 29, 3762–3772 (2010)
Bischler, N. et al. Localization of the yeast RNA polymerase I-specific subunits. EMBO J. 21, 4136–4144 (2002)
Schultz, P., Celia, H., Riva, M., Sentenac, A. & Oudet, P. Three-dimensional model of yeast RNA polymerase I determined by electron microscopy of two-dimensional crystals. EMBO J. 12, 2601–2607 (1993)
Jennebach, S., Herzog, F., Aebersold, R. & Cramer, P. Crosslinking-MS analysis reveals RNA polymerase I domain architecture and basis of rRNA cleavage. Nucleic Acids Res. 40, 5591–5601 (2012)
Fernández-Tornero, C. et al. Insights into transcription initiation and termination from the electron microscopy structure of yeast RNA polymerase III. Mol. Cell 25, 813–823 (2007)
Cramer, P., Bushnell, D. A. & Kornberg, R. D. Structural basis of transcription: RNA polymerase II at 2.8 Ångstrom resolution. Science 292, 1863–1876 (2001)
Karplus, P. A. & Diederichs, K. Linking crystallographic model and data quality. Science 336, 1030–1033 (2012)
Armache, K. J., Kettenberger, H. & Cramer, P. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc. Natl Acad. Sci. USA 100, 6964–6968 (2003)
Bushnell, D. A. & Kornberg, R. D. Complete, 12-subunit RNA polymerase II at 4.1-Å resolution: implications for the initiation of transcription. Proc. Natl Acad. Sci. USA 100, 6969–6973 (2003)
Gnatt, A. L., Cramer, P., Fu, J., Bushnell, D. A. & Kornberg, R. D. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science 292, 1876–1882 (2001)
Kettenberger, H., Armache, K. J. & Cramer, P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell 16, 955–965 (2004)
Hirata, A., Klein, B. J. & Murakami, K. S. The X-ray crystal structure of RNA polymerase from Archaea. Nature 451, 851–854 (2008)
Zhang, G. et al. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell 98, 811–824 (1999)
Vassylyev, D. G. et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417, 712–719 (2002)
Tan, L., Wiesler, S., Trzaska, D., Carney, H. C. & Weinzierl, R. O. Bridge helix and trigger loop perturbations generate superactive RNA polymerases. J. Biol. 7, 40 (2008)
Jovanovic, M. et al. Activity map of the Escherichia coli RNA polymerase bridge helix. J. Biol. Chem. 286, 14469–14479 (2011)
Weinzierl, R. O. The bridge helix of RNA polymerase acts as a central nanomechanical switchboard for coordinating catalysis and substrate movement. Archaea 2011, 608385 (2011)
Edwards, A. M., Kane, C. M., Young, R. A. & Kornberg, R. D. Two dissociable subunits of yeast RNA polymerase II stimulate the initiation of transcription at a promoter in vitro . J. Biol. Chem. 266, 71–75 (1991)
Mosley, A. L. et al. Quantitative proteomics demonstrates that the RNA polymerase II subunits Rpb4 and Rpb7 dissociate during transcription elongation. Mol. Cell. Proteomics 12, 1230–1538 (2013)
Huet, J., Buhler, J. M., Sentenac, A. & Fromageot, P. Dissociation of two polypeptide chains from yeast RNA polymerase A. Proc. Natl Acad. Sci. USA 72, 3034–3038 (1975)
Peyroche, G. et al. The recruitment of RNA polymerase I on rDNA is mediated by the interaction of the A43 subunit with Rrn3. EMBO J. 19, 5473–5482 (2000)
Blattner, C. et al. Molecular basis of Rrn3-regulated RNA polymerase I initiation and cell growth. Genes Dev. 25, 2093–2105 (2011)
Milkereit, P., Schultz, P. & Tschochner, H. Resolution of RNA polymerase I into dimers and monomers and their function in transcription. Biol. Chem. 378, 1433–1443 (1997)
Kettenberger, H., Armache, K. J. & Cramer, P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell 114, 347–357 (2003)
Jeon, C., Yoon, H. & Agarwal, K. The transcription factor TFIIS zinc ribbon dipeptide Asp-Glu is critical for stimulation of elongation and RNA cleavage by RNA polymerase II. Proc. Natl Acad. Sci. USA 91, 9106–9110 (1994)
Prescott, E. M. et al. Transcriptional termination by RNA polymerase I requires the small subunit Rpa12p. Proc. Natl Acad. Sci. USA 101, 6068–6073 (2004)
Kedinger, C., Gniazdowski, M., Mandel, J. L., Jr, Gissinger, F. & Chambon, P. α-Amanitin: a specific inhibitor of one of two DNA-pendent RNA polymerase activities from calf thymus. Biochem. Biophys. Res. Commun. 38, 165–171 (1970)
Weinmann, R. & Roeder, R. G. Role of DNA-dependent RNA polymerase 3 in the transcription of the tRNA and 5S RNA genes. Proc. Natl Acad. Sci. USA 71, 1790–1794 (1974)
De Carlo, S., Carles, C., Riva, M. & Schultz, P. Cryo-negative staining reveals conformational flexibility within yeast RNA polymerase I. J. Mol. Biol. 329, 891–902 (2003)
Chen, Z. A. et al. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29, 717–726 (2010)
Eichner, J., Chen, H. T., Warfield, L. & Hahn, S. Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. EMBO J. 29, 706–716 (2010)
He, Y., Fang, J., Taatjes, D. J. & Nogales, E. Structural visualization of key steps in human transcription initiation. Nature 495, 481–486 (2013)
Beckouet, F. et al. Two RNA polymerase I subunits control the binding and release of Rrn3 during transcription. Mol. Cell. Biol. 28, 1596–1605 (2008)
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010)
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D 59, 2023–2030 (2003)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Skubák, P., Murshudov, G. N. & Pannu, N. S. Direct incorporation of experimental phase information in model refinement. Acta Crystallogr. D 60, 2196–2201 (2004)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Smart, O. S. et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D 68, 368–380 (2012)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Wittekind, M. et al. Isolation and characterization of temperature-sensitive mutations in RPA190, the gene encoding the largest subunit of RNA polymerase I from Saccharomyces cerevisiae . Mol. Cell. Biol. 8, 3997–4008 (1988)
Janke, C. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 (2004)
Ito, H., Fukuda, Y., Murata, K. & Kimura, A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168 (1983)
Acknowledgements
We thank H. Grötsch for preparing the loop deletion yeast strain, and G. von Scheven and A. Scholz for technical assistance. We are also grateful to C. Vonrhein, G. Bricogne, S. Glatt and A. Romero for advice and discussions. We thank staff from the European synchrotrons SOLEIL, DESY, ESRF and SLS, where data were collected during different stages of the project. In particular, we thank A. Thompson for access and support at beamline Proxima 1 (Soleil) and T. Schneider and G. Bourenkov at beamline P14 (PETRA III). We also acknowledge support by the EMBL Heidelberg Protein Expression and Purification, Proteomics Core Facilities and Crystallization Platform, and the ‘Fermentation et culture de microorganisms’ (IFR88, CNRS). We are grateful to M. Bauzan, E. Poilpre and J. Scheurich for yeast fermentation. M.M.-M. and U.J.R. were supported by EMBO Long-Term fellowships, M.M.-M. by the Marie-Curie fellowship (FP7-PEOPLE-2011-IEF 301002), N.M.I.T. by a Fundación Futuro fellowship, F.M.R. by an ESF/CSIC funded JAE-DOC contract and T.G. by the Volkswagen Stiftung via the Niedersachsenprofessur of Prof. G. M. Sheldrick. This work was also partly funded by grant BFU2010-16336 of the Spanish Ministry of Science.
Author information
Authors and Affiliations
Contributions
C.F.-T. and C.W.M. initiated the project. C.F.-T. and U.S. established the Pol I purification and obtained Pol I crystals. C.F.-T., M.M.-M. and U.J.R. further improved the Pol I crystals, and collected data and obtained experimental phase information with the help of N.M.I.T., T.G. and P.L. C.F.-T., M.M.-M., N.M.I.T., F.M.R., T.G. and P.L. carried out the crystallographic analysis and model refinement. C.F.-T., M.M.-M. and C.W.M. wrote the manuscript with input from the other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
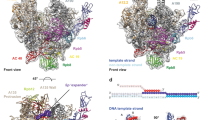
Extended Data Figure 1 Electron densities of different regions of the Pol I structure.
a, Helix in the cleft of subunit A190. Subunits are coloured according to the code given in Fig. 1a. In a–d, σA-weighted electron densities contoured at 1σ are depicted in blue. b, Interaction between subunit Rpb8 (green) and subunit A190 (grey). c, Linker between the two Zn ribbon domains in subunit A12.2. d, A49–A34.5 heterodimer and the anchoring onto the Pol I core by the A34.5 hook and the A49 linker region. e, Anomalous difference Fourier map (purple) calculated from partially selenomethionine-substituted Pol I contoured at 3σ (Extended Data Table 1). In the A49–A34.5 heterodimer two selenium peaks correspond to A34.5 Met 80 and Met 107. f, Anomalous difference Fourier map (purple) showing selenomethionine positions contoured at 3σ. In total, 90 out of 100 expected selenium positions were located within a distance of less than 2.3 Å to corresponding methionine residues.
Extended Data Figure 2 Comparison between Pol I, Pol II and archaeal Pol.
a, Crystal structures of yeast Pol I and Pol II and the archaeal Pol are represented in the same orientation using the same colour code. Whereas the overall organization is conserved, additional subunits such as the A49–A34.5 heterodimer in Pol I and Rpo13 in Archaea are also present. The archaeal Pol lacks the orthologue of subunit A12.2 in Pol I (or Rpb9 in Pol II). The relative position of the stalk also varies between the three RNA polymerases. b, Crystal structures of the individual subunits varying between the three enzymes are depicted. The same colour code for corresponding or identical subunits in the three RNA polymerases is used.
Extended Data Figure 3 Opening of the cleft varies among different Pol I structures and between Pol I and Pol II.
a, Middle panel: front view of the Pol I structure in crystal form C2-100 (see Extended Data Table 1). The complex is divided into two modules. Module 1 (red) is formed by the major part of subunit A190 (without the pore 1, funnel and jaw domains), the C terminus of A135, Rpb5, Rpb6, Rpb8 and the stalk subunits, whereas module 2 (blue) comprises the remaining A135 domains, the pore 1, funnel and jaw domains of A190, AC40–AC19, Rpb10, Rpb12, A12.2 and the A49–A34.5 heterodimer. These modules are held by three hinges in A190 (active site–pore 1 connection, bridge helix and jaw–cleft connection) and one hinge in A135 (hybrid binding–anchor connection) as indicated in Fig. 1b. Pol I structures obtained in crystal forms C2-90 (left panel) and C2-93 (right panel) were superimposed with the one obtained in crystal form C2-100 taking module 2 as reference. Differences between the different crystal forms in the cleft aperture and the tilting of the mobile modules are indicated. b, Pol II structure (Protein Data Bank accession 1WCM) is superimposed onto Pol I (C2-100) using module 1 as reference. In comparison, the Pol II cleft is closed by 10 Å and the modules rotate 15.6°. c, Schematic representation of Pol I and Pol II showing the mobility between Pol I modules, as well as the conformation of the stalk and the clamp. Colour coding is as in b, with the exception of the Pol II stalk, which is coloured deep orange. d, Conformation of the bridge helix of the bacterial Thermus thermophilus polymerase (bacPol, pink, Protein Data Bank accession 1IW7), Pol I (green) and Pol II (yellow, Protein Data Bank accession 1WCM). In addition, the trigger loop is shown for the Thermus thermophilus polymerase, where it is ordered, and as dotted lines for Pol I and Pol II, where it is disordered. e, Sequence alignment of the bridge helix of Saccharomyces cerevisiae and Homo sapiens Pol I, Pol II and Pol III, archaeal Methanococcus jannaschii and Sulfolobus solfataricus, and the bacterial Escherichia coli and Thermus thermophilus polymerases. The secondary structure of the Pol I bridge helix is shown above the alignment. In Methanococcus jannaschii, site-directed mutations Q823D and S824P in subunit A′ lead to increased transcriptional activity.
Extended Data Figure 4 Elongation complex.
a, Cartoon representation of a model of Pol I in complex with an elongation bubble, generated by superposition of the Pol II elongation complex crystal structure (Protein Data Bank accession 1Y1W) using the largest subunit as reference. Whereas Pol II is not shown, the coding and non-coding DNA strands are depicted in blue and cyan, respectively, and the RNA in red. The main Pol I elements putatively involved in nucleic acid interaction appear in different colours, whereas the rest of the Pol I structure is shown in light grey. b, Proposed rearrangements in elongating Pol I (coloured elements) in analogy with Pol II (grey elements). Closure of the cleft is expected to approach the wall (tan), the hybrid binding domain (dark red) and the fork loop 2 (orange) to the bubble, as well as to fold the bridge helix (green). Closure of the fork loop 1 (pink) and opening of the lid loop (cyan) would also be required.
Extended Data Figure 5 The DNA-mimicking loop of Pol I forms a mobile element.
a, σA-weighted electron density contoured at 1σ of A190 jaw residues 1361–1399 (middle and right panel) in crystal form C2-90. Density is also present in crystal form C2-93 (data not shown), whereas it is absent in crystal form C2-100 at the same contour level (left panel). b, Sequence alignment of the Pol I DNA-mimicking loop across different species highlighting the conservation of this element. c, Purified Pol I shows elongation activity in an RNA extension assay. DNA templates (Temp-41 and Temp-27 of 41 and 27 nucleotides, respectively) and 32P-labelled RNA sequences used for the assay are indicated. The autoradiogram shows the elongation of RNA by Pol I producing a run-off of 18 nucleotides (lane 3) or 12 nucleotides (lane 6) depending of the template used. Lanes 1 and 4: the DNA/RNA hybrids were incubated in the absence of Pol I. Lanes 2 and 5: Pol I–DNA–RNA complexes were incubated with a buffer without NTPs. d, Dot spots grown at the indicated temperatures of the parental RPA190 strain and rpa190Δloop strain where the DNA-mimicking loop has been deleted. The rpa190Δloop strain shows a slight temperature-sensitive growth defect on SDC medium.
Extended Data Figure 6 Pol I dimer in the crystal lattice.
The A43 C-terminal tail establishes crystal contacts with a second molecule related by a crystallographic dyad. The A43 C-terminal helix is embedded between the clamp and the protrusion domain of a dyad related molecule. The σA-weighted electron density map (contoured at 1σ) shows clear density corresponding to residues A43 251–326. The two monomers are related by a crystallographic dyad, which is indicated by a dyad symbol.
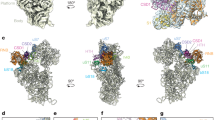
Extended Data Figure 7 Subunit A12.2 structure and its position in Pol I.
a, Detailed views show the A12.2 Zn sites and the main contacts between its linker and the A190 subunit. The A12.2 linker extends the β-sheet of the A190 jaw. b, The overlap between subunit A12.2 and α-amanitin in the Pol I structure explains the insensitivity of Pol I for this fungal toxin. The Pol II–α-amanitin complex structure (Protein Data Bank accession 2VUM) was superimposed onto the Pol I crystal structure. In the left panel, the α-amanitin toxin is depicted in surface representation (pink). On the right, a detailed view of α-amanitin shows the overlap with the C-terminal Zn ribbon of A12.2.
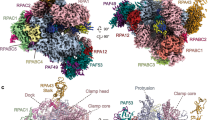
Extended Data Figure 8 Precise positioning of the A49–A34.5 heterodimer suggests similar positions for the related C37–C53 heterodimer in Pol III and the TFIIF heterodimer in Pol II.
a, Pol I was fitted into the Pol III envelope (EM-1804)12. A49–A34.5 (corresponding to C37–C53 in Pol III), AC40–AC19 and A12.2 (corresponding to C11 in Pol III) are coloured as in Fig. 1. The approximate position of subcomplex C82–C34–C31 is also indicated. b, The proposed Pol II/TFIIF model was manually fitted into the Pol II/TFII-A-B-F/TBP/DNA EM density (EM-2305)43. c, Left panel: detailed view of the anchoring of the A49–A34.5 dimerization domain onto the Pol I core. Right panel: model for the TFIIF dimerization module bound to the Pol II core based on the crystal structures of the human Rap74–Rap30 complex (Protein Data Bank accession 1F3U) and Pol II (Protein Data Bank accession 1WCM).
Rights and permissions
About this article
Cite this article
Fernández-Tornero, C., Moreno-Morcillo, M., Rashid, U. et al. Crystal structure of the 14-subunit RNA polymerase I. Nature 502, 644–649 (2013). https://doi.org/10.1038/nature12636
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature12636