Abstract
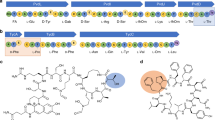
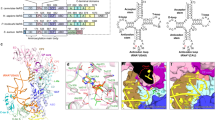
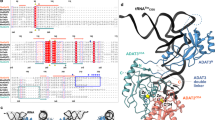
Mistranslation arising from confusion of serine for alanine by alanyl-tRNA synthetases (AlaRSs) has profound functional consequences1,2,3. Throughout evolution, two editing checkpoints prevent disease-causing mistranslation from confusing glycine or serine for alanine at the active site of AlaRS. In both bacteria and mice, Ser poses a bigger challenge than Gly1,2. One checkpoint is the AlaRS editing centre, and the other is from widely distributed AlaXps—free-standing, genome-encoded editing proteins that clear Ser-tRNAAla. The paradox of misincorporating both a smaller (glycine) and a larger (serine) amino acid suggests a deep conflict for nature-designed AlaRS. Here we show the chemical basis for this conflict. Nine crystal structures, together with kinetic and mutational analysis, provided snapshots of adenylate formation for each amino acid. An inherent dilemma is posed by constraints of a structural design that pins down the α-amino group of the bound amino acid by using an acidic residue. This design, dating back more than 3 billion years, creates a serendipitous interaction with the serine OH that is difficult to avoid. Apparently because no better architecture for the recognition of alanine could be found, the serine misactivation problem was solved through free-standing AlaXps, which appeared contemporaneously with early AlaRSs. The results reveal unconventional problems and solutions arising from the historical design of the protein synthesis machinery.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Protein Data Bank
Data deposits
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession codes 3hxu (WT/Ala-SA), 3hxv (WT/Gly-SA), 3hxw (WT/Ser-SA), 3hxx (WT/AMP-PCP/Mg(II)), 3hxy (WT/Ala-AMP/PCP/AMP-PCP/Mg(II)), 3hxz (G237A/Ala-SA), 3hy0 (G237A/Gly-SA) and 3hy1 (G237A-apo and G237A/Ser-SA).
References
Beebe, K., Ribas De Pouplana, L. & Schimmel, P. Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability. EMBO J. 22, 668–675 (2003)
Lee, J. W. et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 443, 50–55 (2006)
Beebe, K., Mock, M., Merriman, E. & Schimmel, P. Distinct domains of tRNA synthetase recognize the same base pair. Nature 451, 90–93 (2008)
Carter, C. W. Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu. Rev. Biochem. 62, 715–748 (1993)
Giege, R. The early history of tRNA recognition by aminoacyl-tRNA synthetases. J. Biosci. 31, 477–488 (2006)
Norris, A. T. & Berg, P. Mechanism of aminoacyl RNA synthesis: studies with isolated aminoacyl adenylate complexes of isoleucyl RNA synthetase. Proc. Natl Acad. Sci. USA 52, 330–337 (1964)
Eldred, E. W. & Schimmel, P. R. Rapid deacylation by isoleucyl transfer ribonucleic acid synthetase of isoleucine-specific transfer ribonucleic acid aminoacylated with valine. J. Biol. Chem. 247, 2961–2964 (1972)
Boniecki, M. T., Vu, M. T., Betha, A. K. & Martinis, S. A. CP1-dependent partitioning of pretransfer and posttransfer editing in leucyl-tRNA synthetase. Proc. Natl Acad. Sci. USA 105, 19223–19228 (2008)
Fersht, A. R. Enzyme Structure and Mechanism (Freeman, 1977)
Nureki, O. et al. Structural basis for amino acid and tRNA recognition by class I aminoacyl-tRNA synthetases. Cold Spring Harb. Symp. Quant. Biol. 66, 167–173 (2001)
Fersht, A. R. Sieves in sequence. Science 280, 541 (1998)
Fukai, S. et al. Structural basis for double-sieve discrimination of l-valine from l-isoleucine and l-threonine by the complex of tRNAVal and valyl-tRNA synthetase. Cell 103, 793–803 (2000)
Tsui, W. C. & Fersht, A. R. Probing the principles of amino acid selection using the alanyl-tRNA synthetase from Escherichia coli . Nucleic Acids Res. 9, 4627–4637 (1981)
Ahel, I., Korencic, D., Ibba, M. & Söll, D. Trans-editing of mischarged tRNAs. Proc. Natl Acad. Sci. USA 100, 15422–15427 (2003)
Chong, Y. E., Yang, X. L. & Schimmel, P. Natural homolog of tRNA synthetase editing domain rescues conditional lethality caused by mistranslation. J. Biol. Chem. 283, 30073–30078 (2008)
Ling, J. et al. Resampling and editing of mischarged tRNA prior to translation elongation. Mol. Cell 33, 654–660 (2009)
Guo, M. et al. The C-Ala domain brings together editing and aminoacylation functions on one tRNA. Science 325, 744–747 (2009)
Sokabe, M., Okada, A., Yao, M., Nakashima, T. & Tanaka, I. Molecular basis of alanine discrimination in editing site. Proc. Natl Acad. Sci. USA 102, 11669–11674 (2005)
Ho, B. K. & Gruswitz, F. HOLLOW: generating accurate representations of channel and interior surfaces in molecular structures. BMC Struct. Biol. 8, 49 (2008)
Arnez, J. G. & Moras, D. Structural and functional considerations of the aminoacylation reaction. Trends Biochem. Sci. 22, 211–216 (1997)
Davis, M. W., Buechter, D. D. & Schimmel, P. Functional dissection of a predicted class-defining motif in a class II tRNA synthetase of unknown structure. Biochemistry 33, 9904–9911 (1994)
Jakubowski, H. in The Aminoacyl-tRNA Synthetases (eds Ibba, M., Francklyn, C. & Cusack, S.) 384–396 (Eurekah, 2005)
Shi, J. P., Musier-Forsyth, K. & Schimmel, P. Region of a conserved sequence motif in a class II tRNA synthetase needed for transfer of an activated amino acid to an RNA substrate. Biochemistry 33, 5312–5318 (1994)
Xin, Y., Li, W. & First, E. A. Stabilization of the transition state for the transfer of tyrosine to tRNATyr by tyrosyl-tRNA synthetase. J. Mol. Biol. 303, 299–310 (2000)
Sankaranarayanan, R. et al. Zinc ion mediated amino acid discrimination by threonyl-tRNA synthetase. Nature Struct. Biol. 7, 461–465 (2000)
First, E. A. in The Aminoacyl-tRNA Synthetases (eds Ibba, M., Francklyn, C. & Cusack, S.) 328–352 (Eurekah, 2005)
Belrhali, H. et al. Crystal structures at 2.5 angstrom resolution of seryl-tRNA synthetase complexed with two analogs of seryl adenylate. Science 263, 1432–1436 (1994)
Cieslik, M. & Derewenda, Z. S. The role of entropy and polarity in intermolecular contacts in protein crystals. Acta Crystallogr. D 65, 500–509 (2009)
Itoh, Y. et al. Crystallographic and mutational studies of seryl-tRNA synthetase from the archaeon Pyrococcus horikoshii . RNA Biol. 5, 169–177 (2008)
Swairjo, M. A. & Schimmel, P. R. Breaking sieve for steric exclusion of a noncognate amino acid from active site of a tRNA synthetase. Proc. Natl Acad. Sci. USA 102, 988–993 (2005)
Malde, A. K. & Mark, A. E. Binding and enantiomeric selectivity of threonyl-tRNA synthetase. J. Am. Chem. Soc. 131, 3848–3849 (2009)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Acknowledgements
We thank R. J. Read for pointing out the errors in the previous AlaRS–ligand structures and for crystallographic discussions; and G. J. Kleywegt, Z. Otwinowski and A. Perrakis for technical assistance. X-ray diffraction data were collected at Stanford Synchrotron Radiation Laboratory (SSRL) beamlines 7-1, 9-1 and 11-1. This work was supported by grant GM 15539 from the National Institutes of Health and by a fellowship from the National Foundation for Cancer Research.
Author Contributions M.G., X.-L.Y. and P.S. designed the experiments. M.G., Y.E.C., R.S. and K.B. performed the experiments. M.G. and Y.E.C. analysed the data. M.G., Y.E.C., X.-L.Y. and P.S. wrote the paper. All authors discussed the results and commented on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-8 with Legends. (PDF 5485 kb)
Supplementary Table 1
This file contains a summary of the data collection and refinement statistics of the nine structures. (PDF 192 kb)
Supplementary Movie 1
This movie file shows that serine paradox is caused by AlaRS recognition dilemma (see Supplementary Figures file for full Legend). (MOV 6540 kb)
Supplementary Movie 2
This movie file shows the alanyl-adenylate formation mechanism (see Supplementary Figures file for full Legend). (MOV 4436 kb)
Rights and permissions
About this article
Cite this article
Guo, M., Chong, Y., Shapiro, R. et al. Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma. Nature 462, 808–812 (2009). https://doi.org/10.1038/nature08612
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature08612
This article is cited by
-
The structural basis of the genetic code: amino acid recognition by aminoacyl-tRNA synthetases
Scientific Reports (2020)
-
ANKRD16 prevents neuron loss caused by an editing-defective tRNA synthetase
Nature (2018)
-
Emerging roles of tRNA in adaptive translation, signalling dynamics and disease
Nature Reviews Genetics (2015)
-
The selective tRNA aminoacylation mechanism based on a single G•U pair
Nature (2014)
-
Guanidine Hydrochloride Mediated Denaturation of E. coli Alanyl-tRNA Synthetase: Identification of an Inactive Dimeric Intermediate
The Protein Journal (2014)