Abstract
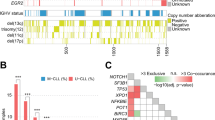
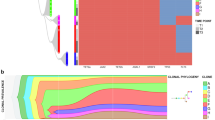
High-count monoclonal B-cell lymphocytosis (MBL) is an asymptomatic expansion of clonal B cells in the peripheral blood without other manifestations of chronic lymphocytic leukemia (CLL). Yearly, 1% of MBLs evolve to CLL requiring therapy; thus being critical to understand the biological events that determine which MBLs progress to intermediate/advanced CLL. In this study, we performed targeted deep sequencing on 48 high-count MBLs, 47 of them with 2–4 sequential samples analyzed, exploring the mutation status of 21 driver genes and evaluating clonal evolution. We found somatic non-synonymous mutations in 25 MBLs (52%) at the initial time point analyzed, including 12 (25%) with >1 mutated gene. In cases that subsequently progressed to CLL, mutations were detected 41 months (median) prior to progression. Excepting NOTCH1, TP53 and XPO1, which showed a lower incidence in MBL, genes were mutated with a similar prevalence to CLL, indicating the early origin of most driver mutations in the MBL/CLL continuum. MBLs with mutations at the initial time point analyzed were associated with shorter time-to-treatment (TTT). Furthermore, MBLs showing subclonal expansion of driver mutations on sequential evaluation had shorter progression time to CLL and shorter TTT. These findings support that clonal evolution has prognostic implications already at the pre-malignant MBL stage, anticipating which individuals will progress earlier to CLL.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Call TG, Phyliky RL, Noel P, Habermann TM, Beard CM, O'Fallon WM et al. Incidence of chronic lymphocytic leukemia in Olmsted County, Minnesota, 1935 through 1989, with emphasis on changes in initial stage at diagnosis. Mayo Clin Proc 1994; 69: 323–328.
Diehl LF, Karnell LH, Menck HR . The American College of Surgeons Commission on Cancer and the American Cancer Society. The National Cancer Data Base report on age, gender, treatment, and outcomes of patients with chronic lymphocytic leukemia. Cancer 1999; 86: 2684–2692.
Baliakas P, Hadzidimitriou A, Sutton LA, Rossi D, Minga E, Villamor N et al. Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia 2015; 29: 329–336.
Stilgenbauer S, Schnaiter A, Paschka P, Zenz T, Rossi M, Dohner K et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood 2014; 123: 3247–3254.
Rossi D, Bruscaggin A, Spina V, Rasi S, Khiabanian H, Messina M et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine-refractoriness. Blood 2011; 118: 6904–6908.
Jeromin S, Weissmann S, Haferlach C, Dicker F, Bayer K, Grossmann V et al. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia 2014; 28: 108–117.
Pflug N, Bahlo J, Shanafelt TD, Eichhorst BF, Bergmann MA, Elter T et al. Development of a comprehensive prognostic index for patients with chronic lymphocytic leukemia. Blood 2014; 124: 49–62.
Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell 2013; 152: 714–726.
Quesada V, Conde L, Villamor N, Ordonez GR, Jares P, Bassaganyas L et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet 2012; 44: 47–52.
Schuh A, Becq J, Humphray S, Alexa A, Burns A, Clifford R et al. Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood 2012; 120: 4191–4196.
Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med 2011; 365: 2497–2506.
Puente XS, Bea S, Valdes-Mas R, Villamor N, Gutierrez-Abril J, Martin-Subero JI et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature 2015; 526: 519–524.
Ojha J, Secreto CR, Rabe KG, Van Dyke DL, Kortum KM, Slager SL et al. Identification of recurrent truncated DDX3X mutations in chronic lymphocytic leukaemia. Br J Haematol 2015; 169: 445–448.
Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J et al. Mutations driving CLL and their evolution in progression and relapse. Nature 2015; 526: 525–530.
Knight SJ, Yau C, Clifford R, Timbs AT, Sadighi Akha E, Dreau HM et al. Quantification of subclonal distributions of recurrent genomic aberrations in paired pre-treatment and relapse samples from patients with B-cell chronic lymphocytic leukemia. Leukemia 2012; 26: 1564–1575.
Braggio E, Kay NE, Vanwier S, Tschumper RC, Smoley S, Eckel-Passow JE et al. Longitudinal genome-wide analysis of patients with chronic lymphocytic leukemia reveals complex evolution of clonal architecture at disease progression and at the time of relapse. Leukemia 2012; 26: 1698–1701.
Ojha J, Ayres J, Secreto C, Tschumper R, Rabe K, Van Dyke D et al. Deep sequencing identifies genetic heterogeneity and recurrent convergent evolution in chronic lymphocytic leukemia. Blood 2015; 125: 492–498.
Marti GE, Rawstron AC, Ghia P, Hillmen P, Houlston RS, Kay N et al. Diagnostic criteria for monoclonal B-cell lymphocytosis. Br J Haematol 2005; 130: 325–332.
Shanafelt TD, Ghia P, Lanasa MC, Landgren O, Rawstron AC . Monoclonal B-cell lymphocytosis (MBL): biology, natural history and clinical management. Leukemia 2010; 24: 512–520.
Rawstron AC, Bennett FL, O'Connor SJ, Kwok M, Fenton JA, Plummer M et al. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N Engl J Med 2008; 359: 575–583.
Greco M, Capello D, Bruscaggin A, Spina V, Rasi S, Monti S et al. Analysis of SF3B1 mutations in monoclonal B-cell lymphocytosis. Hematol Oncol 2013; 31: 54–55.
Rasi S, Monti S, Spina V, Foa R, Gaidano G, Rossi D . Analysis of NOTCH1 mutations in monoclonal B-cell lymphocytosis. Haematologica 2012; 97: 153–154.
Ojha J, Secreto C, Rabe K, Ayres-Silva J, Tschumper R, Dyke DV et al. Monoclonal B-cell lymphocytosis is characterized by mutations in CLL putative driver genes and clonal heterogeneity many years before disease progression. Leukemia 2014; 28: 2395–2398.
Lionetti M, Fabris S, Cutrona G, Agnelli L, Ciardullo C, Matis S et al. High-throughput sequencing for the identification of NOTCH1 mutations in early stage chronic lymphocytic leukaemia: biological and clinical implications. Br J Haematol 2014; 165: 629–639.
Winkelmann N, Rose-Zerilli M, Forster J, Parry M, Parker A, Gardiner A et al. Low frequency mutations independently predict poor treatment free survival in early stage chronic lymphocytic leukemia and monoclonal B-cell lymphocytosis. Haematologica 2015; 100: e237–e239.
Shanafelt TD, Kay NE, Rabe KG, Call TG, Zent CS, Maddocks K et al. Brief report: natural history of individuals with clinically recognized monoclonal B-cell lymphocytosis compared with patients with Rai 0 chronic lymphocytic leukemia. J Clin Oncol 2009; 27: 3959–3963.
Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M et al. An integrated semiconductor device enabling non-optical genome sequencing. Nature 2011; 475: 348–352.
Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011; 475: 101–105.
Kocher JP, Quest DJ, Duffy P, Meiners MA, Moore RM, Rider D et al. The Biological Reference Repository (BioR): a rapid and flexible system for genomics annotation. Bioinformatics 2014; 30: 1920–1922.
Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G et al. Integrative genomics viewer. Nat Biotechnol 2011; 29: 24–26.
Hallek M, Cheson BD, Catovsky D, Caligaris-Cappio F, Dighiero G, Dohner H et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood 2008; 111: 5446–5456.
Ouillette P, Saiya-Cork K, Seymour E, Li C, Shedden K, Malek SN . Clonal evolution, genomic drivers, and effects of therapy in chronic lymphocytic leukemia. Clin Cancer Res 2013; 19: 2893–2904.
Guieze R, Robbe P, Clifford R, de Guibert S, Pereira B, Timbs A et al. Presence of multiple recurrent mutations revealed by targeted NGS confers poor trial outcome of relapsed/refractory CLL. Blood 2015; 126: 2110–2117.
Rossi D, Khiabanian H, Spina V, Ciardullo C, Bruscaggin A, Fama R et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood 2014; 123: 2139–2147.
Acknowledgements
This work was supported by the Henry Predolin Foundation and NIH grant CA197120.
Author contributions
SB, JO, CS, KMK, SP and EB performed the experiments; SB, KGC, SLS and EB analyzed the results; SB, TDS, DLV, RF, SLS, NEK and EB designed the research; SB and EB wrote the paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
RF has received a patent for the prognostication of MM based on genetic categorization of the disease. He has received consulting fees from Celgene, Genzyme, BMS, Bayer, Lilly, Onyx, Binding Site, Novartis, Sanofi, Millennium and AMGEN. He also has sponsored research from Onyx. He is also a member of the Scientific Advisory Board of Applied Biosciences.
Additional information
Supplementary Information accompanies this paper on the Leukemia website
Rights and permissions
About this article
Cite this article
Barrio, S., Shanafelt, T., Ojha, J. et al. Genomic characterization of high-count MBL cases indicates that early detection of driver mutations and subclonal expansion are predictors of adverse clinical outcome. Leukemia 31, 170–176 (2017). https://doi.org/10.1038/leu.2016.172
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/leu.2016.172
This article is cited by
-
Lymphoid clonal hematopoiesis: implications for malignancy, immunity, and treatment
Blood Cancer Journal (2023)
-
Chronic lymphocytic leukaemia: from genetics to treatment
Nature Reviews Clinical Oncology (2019)
-
Spectrum and functional validation of PSMB5 mutations in multiple myeloma
Leukemia (2019)
-
Growth dynamics in naturally progressing chronic lymphocytic leukaemia
Nature (2019)
-
Evolving DNA methylation and gene expression markers of B-cell chronic lymphocytic leukemia are present in pre-diagnostic blood samples more than 10 years prior to diagnosis
BMC Genomics (2017)