Abstract
Multiple mutations in SOX2 have been identified in patients with ocular anomalies and/or pituitary dysfunction. Here, we identified SOX2 abnormalities in nine patients. The molecular defects included one missense, one nonsense and four frameshift mutations, and three submicroscopic deletions involving SOX2. Three of the six mutations and all deletions were hitherto unreported. The breakpoints determined in one deletion were located within Alu repeats and accompanied by an overlap of 11 bp. Three of the six mutations encoded SOX2 proteins that lacked in vitro transactivation activity for the HESX1 promoter, whereas the remaining three generated proteins with ∼15–∼20% of transactivation activity. All cases manifested ocular anomalies of various severities, together with several complications including arachnoid cyst and hamartoma. There was no apparent correlation between the residual activity and clinical severity. The results indicate that molecular defects in SOX2 are highly variable and include Alu repeat-mediated genomic rearrangements. Our data provide further evidence for wide phenotypic variation of SOX2 abnormalities and the lack of genotype–phenotype correlation in patients carrying SOX2 lesions.
Similar content being viewed by others
Main
SOX2 (NP_003097.1) has a critical role in the development of the eye, pituitary and central nervous system through transactivation of multiple genes including HESX1.1, 2, 3 Haploinsufficiency of SOX2 (NM_003106.3) leads to anophthalmia/microphthalmia and pituitary dysfunction, in addition to various neuronal defects such as mental retardation, brain malformation and hearing loss.3, 4, 5, 6, 7 Although >80 patients with SOX2 abnormalities have been reported to date,3, 4, 5, 6, 7, 8, 9, 10, 11 current understanding of mutation spectrum and phenotypic determinants of this condition remains fragmentary. In fact, in vitro functional assays have been performed only for a small number of mutations.3, 11, 12, 13 Furthermore, whereas Kelberman et al.12 found no obvious genotype–phenotype correlation, Schneider et al.14 reported that patients with missense mutations had milder ocular phenotypes than those with nonsense or frameshift mutations.
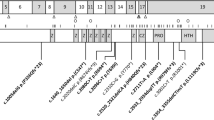
Here, we identified SOX2 abnormalities in nine patients (cases 1–9). This study was approved by the Institutional Review Board Committee at the National Center for Child Health and Development and performed after obtaining written informed consent. Molecular defects in cases 1–9 were identified through sequencing and copy number analyses of SOX2 for 37 patients with ocular anomalies and 15 patients with pituitary dysfunction and normal eyes (Supplementary Table S1). Mutations in the coding region were examined by direct sequencing, and copy number abnormalities were analyzed by multiplex ligation-dependent amplification and array comparative genomic hybridization. Detailed methods are provided in Supplementary Table S2. Cases 1–6 carried heterozygous intragenic mutations, whereas cases 7–9 had heterozygous submicroscopic deletions involving SOX2. Cases 1–9 invariably manifested developmental defects of eyes, in addition to multiple complications including arachnoid cyst and hamartoma (Table 1 and Supplementary Table S3). The ocular phenotypes included unilateral and bilateral microphthalmia, bilateral coloboma and bilateral anophthalmia. The SOX2 mutations in cases 1–6 consisted of three previously reported mutations (c.70_89del20, c.70_86del17 and c.480C>G) and three novel mutations (c.235 T>C, c.244_245delTT and c.402delC) (Figure 1a). None of the six mutations have been registered as polymorphisms in the single-nucleotide polymorphism database (dbSNP, http://www.ncbi.nlm.nih.gov/). In vitro reporter assays using a vector containing the HESX1 promoter indicated that c.235T>C, c.402delC and c.480C>G encoded proteins with ∼15–∼20% transactivation activity, whereas the remaining three generated proteins that lacked the activity (Figure 1b). Of the six mutations, c.70_89del20, c.70_86del17 and c.244_245delTT affected the possible nuclear localization signals predicted from mouse data, and therefore seemed to disturb the intracellular localization of the SOX2 protein (Figure 1a).15 Furthermore, it remained possible that mRNAs of the nonsense and frameshift mutations undergo early degradation in vivo. The deletions in cases 7–9 affected ∼1.0 to ∼2.5 Mb genomic regions at 3q26-27 including SOX2. These deletions overlapped with, but were not identical to, previously reported deletions (Figure 2).4, 7, 8, 9, 12, 13, 14, 16 Sequences at the fusion junction were characterized in case 7, showing that the two breakpoints resided within Alu repeats and shared an overlap of 11 bp (Figure 2).
The position and in vitro function of SOX2 mutations identified in the present study. (a) Chromatograms of the mutations. Subcloned wild-type (WT) and mutant (MT) sequences are shown. Deleted nucleotides are shaded in gray and mutated nucleotides are indicated by asterisks. The yellow boxes indicate the position of two putative nuclear localization signals (NLSs) predicted from mouse data.15 (b) In vitro reporter assay using a luciferase vector containing the HESX1 promoter. The results are expressed as the mean±1 s.d. Asterisks indicate the statistical significance of the results (P⩽0.05). The relative fold activation of c.70_89del20, c.70_86del17 and c.244_245delTT was similar to that of the empty vector (empty). The relative percentages of fold activation of c.235T>C, c.402delC and c.480C>G to that of the WT SOX2 were 13.8%, 15.4% and, 18.4%, respectively.
Submicroscopic deletions identified in the present and previous studies. Upper panel: genomic structure of the 3q26-27 region and the position of previously reported deletions. The position of SOX2 is indicated by a red box and the deletions are depicted by black bars. Genomic positions of the genes and deletions refer to NCBI database (http://www.ncbi.nlm.nih.gov/). Cen, centromere; tel, telomere. Lower panel: comparative genomic hybridization analysis of cases 7–9 from the present study and the sequence at the fusion junction in case 7. The black, red and green dots denote signals indicative of the normal, increased (>+0.5) and decreased (<−1.0) copy numbers, respectively. Estimated sizes and positions of the heterozygous deletions are shown.
Several matters are noteworthy. First, molecular lesions in cases 1–9 were heterogeneous and included six point mutations and three submicroscopic deletions. These data support a broad mutation spectrum of SOX2 abnormalities.8, 9 Notably, we identified submicroscopic deletions involving SOX2 in three cases. As multiple microdeletions at 3q26-27 have been reported in patients with ocular anomalies (Figure 2),4, 7, 8, 9, 12, 13, 14, 16 it is possible that the genomic region around SOX2 represents a hotspot for chromosomal rearrangements. In this regard, it is noteworthy that the sequence at the fusion junction in case 7 is consistent with non-allelic homologous recombination that occurs between two homologous sequences or replication-based errors that are usually associated with microhomology at the fusion junction.17 Furthermore, the two breakpoints of this deletion resided within Alu repeats, which facilitate both recombination- and replication-mediated errors.18, 19 These data imply that Alu repeats may have a role in the high frequency of deletions at 3q26-27. Second, cases 1–9 manifested several complications in addition to ocular abnormalities. Importantly, seven cases manifested brain anomalies including arachnoid cyst and hamartoma. In this regard, Alatzoglou et al.13 recently described pituitary tumors in two patients with SOX2 haploinsufficiency. Thus, SOX2 abnormalities seem to be associated with various types of developmental defects and tumors in the brain. Third, SOX2 lesions were identified in 9 of 37 patients with ocular abnormalities, but were absent from 15 patients with pituitary dysfunction and normal eyes. The results are consistent with the previous reports that SOX2 abnormalities account for 10–20% of the etiology of anophthalmia/microphthalmia and rarely result in pituitary dysfunction without eye abnormalities.4, 5, 8, 9, 11 These data can be explained by assuming that, during development, the eye is highly sensitive to reduced activity of SOX2. Nevertheless, as gonadotropin deficiency was observed in all of our mutation-positive cases, SOX2 appears to be indispensable for the function of the hypothalamus–pituitary axis. Lastly, no apparent genotype–phenotype correlation was found in our patient cohort. Although ocular phenotypes were relatively mild in cases 1 and 6 with hypomorphic mutations and obviously severe in cases 3 and 4 with amorphic mutations, bilateral anophthalmia was also observed in case 2 with a hypomorphic mutation and mild coloboma with normal visual activity was seen in case 9 with SOX2 deletion. Similarly, the occurrence of pituitary dysfunction, mental retardation and short stature was not associated with the mutation types. The lack of correlation between residual activity and phenotypic severity is consistent with the previously proposed notion that haploinsufficiency of developmental genes is usually associated with a wide range of penetrance and expressivity.20
Collectively, the present study argues for a broad spectrum of SOX2 lesions and indicates for the first time that Alu repeat-mediated genomic rearrangements at 3q26-27 account for a part of the etiology of SOX2 haploinsufficiency. Our data provide further evidence for the wide phenotypic variation and the lack of genotype–phenotype correlation in patients with SOX2 abnormalities.
References
Kelberman, D., Rizzoti, K., Lovell-Badge, R., Robinson, I. C. & Dattani, M. T. Genetic regulation of pituitary gland development in human and mouse. Endocr. Rev. 30, 790–829 (2009).
Jayakody, S. A., Andoniadou, C. L., Gaston-Massuet, C., Signore, M., Cariboni, A., Bouloux, P. M. et al. SOX2 regulates the hypothalamic-pituitary axis at multiple levels. J. Clin. Invest. 122, 3635–3646 (2012).
Kelberman, D., Rizzoti, K., Avilion, A., Bitner-Glindzicz, M., Cianfarani, S., Collins, J. et al. Mutations within Sox2/SOX2 are associated with abnormalities in the hypothalamo-pituitary-gonadal axis in mice and humans. J. Clin. Invest. 116, 2442–2455 (2006).
Fantes, J., Ragge, N. K., Lynch, S. A., McGill, N. I., Collin, J. R., Howard-Peebles, P. N. et al. Mutations in SOX2 cause anophthalmia. Nat. Genet. 33, 461–463 (2003).
Stark, Z., Storen, R., Bennetts, B., Savarirayan, R. & Jamieson, R. V. Isolated hypogonadotropic hypogonadism with SOX2 mutation and anophthalmia/microphthalmia in offspring. Eur. J. Hum. Genet. 19, 753–756 (2011).
Hagstrom, S. A., Pauer, G. J., Reid, J., Simpson, E., Crowe, S., Maumenee, I. H. et al. SOX2 mutation causes anophthalmia, hearing loss, and brain anomalies. Am. J. Med. Genet. A 138A, 95–98 (2005).
Williamson, K. A., Hever, A. M., Rainger, J., Rogers, R. C., Magee, A., Fiedler, Z. et al. Mutations in SOX2 cause anophthalmia-esophageal-genital (AEG) syndrome. Hum. Mol. Genet. 15, 1413–1422 (2006).
Bakrania, P., Robinson, D. O., Bunyan, D. J., Salt, A., Martin, A., Crolla, J. A. et al. SOX2 anophthalmia syndrome: 12 new cases demonstrating broader phenotype and high frequency of large gene deletions. Br. J. Ophthalmol. 91, 1471–1476 (2007).
Chassaing, N., Causse, A., Vigouroux, A., Delahaye, A., Alessandri, J. L., Boespflug-Tanguy, O. et al. Molecular findings and clinical data in a cohort of 150 patients with anophthalmia/microphthalmia. Clin. Genet. e-pub ahead of print 10 September 2013 doi:10.1111/cge.12275.
Schilter, K. F., Reis, L. M., Schneider, A., Bardakjian, T. M., Abdul-Rahman, O., Kozel, B. A. et al. Whole-genome copy number variation analysis in anophthalmia and microphthalmia. Clin. Genet. 84, 473–481 (2013).
Sato, N., Kamachi, Y., Kondoh, H., Shima, Y., Morohashi, K., Horikawa, R. et al. Hypogonadotropic hypogonadism in an adult female with a heterozygous hypomorphic mutation of SOX2. Eur. J. Endocrinol. 156, 167–171 (2007).
Kelberman, D., de Castro, S. C., Huang, S., Crolla, J. A., Palmer, R., Gregory, J. W. et al. SOX2 plays a critical role in the pituitary, forebrain, and eye during human embryonic development. J. Clin. Endocrinol. Metab. 93, 1865–1873 (2008).
Alatzoglou, K. S., Andoniadou, C. L., Kelberman, D., Buchanan, C. R., Crolla, J., Arriazu, M. C. et al. SOX2 haploinsufficiency is associated with slow progressing hypothalamo-pituitary tumours. Hum. Mutat. 32, 1376–1380 (2011).
Schneider, A., Bardakjian, T., Reis, L. M., Tyler, R. C. & Semina, E. V. Novel SOX2 mutations and genotype-phenotype correlation in anophthalmia and microphthalmia. Am. J. Med. Genet. A 149A, 2706–2715 (2009).
Li, J., Pan, G., Cui, K., Liu, Y., Xu, S. & Pei, D. A dominant-negative form of mouse SOX2 induces trophectoderm differentiation and progressive polyploidy in mouse embryonic stem cells. J. Biol. Chem. 282, 19481–19492 (2007).
Salem, N. J., Hempel, M., Heiliger, K. J., Hosie, S., Meitinger, T. & Oexle, K. Anal atresia, coloboma, microphthalmia, and nasal skin tag in a female patient with 3.5 Mb deletion of 3q26 encompassing SOX2. Am. J. Med. Genet. A 161A, 1421–1424 (2013).
Gu, W., Zhang, F. & Lupski, J. R. Mechanisms for human genomic rearrangements. Pathogenetics 1, 4 (2008).
Chen, J. M., Cooper, D. N., Férec, C., Kehrer-Sawatzki, H. & Patrinos, G. P. Genomic rearrangements in inherited disease and cancer. Semin. Cancer Biol. 20, 222–233 (2010).
Boone, P. M., Liu, P., Zhang, F., Carvalho, C. M., Towne, C. F., Batish, S. D. et al. Alu-specific microhomology-mediated deletion of the final exon of SPAST in three unrelated subjects with hereditary spastic paraplegia. Genet. Med. 13, 582–592 (2011).
Fisher, E. & Scambler, P. Human haploinsufficiency—one for sorrow, two for joy. Nat. Genet. 7, 5–7 (1994).
Acknowledgements
This work was supported by grants from the Ministry of Health, Labor and Welfare and from Takeda Science Foundation, by Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science, by Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Sports, Science and Technology and by the Grant of National Center for Child Health and Development.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Suzuki, J., Azuma, N., Dateki, S. et al. Mutation spectrum and phenotypic variation in nine patients with SOX2 abnormalities. J Hum Genet 59, 353–356 (2014). https://doi.org/10.1038/jhg.2014.34
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/jhg.2014.34
Keywords
This article is cited by
-
Evaluation of somatic and/or germline mosaicism in congenital malformation of the eye
European Journal of Human Genetics (2023)
-
Genetics of anophthalmia and microphthalmia. Part 2: Syndromes associated with anophthalmia–microphthalmia
Human Genetics (2019)