Abstract
Heterozygous COL2A1 variants cause a wide spectrum of skeletal dysplasia termed type II collagenopathies. We assessed the impact of this gene in our French series. A decision tree was applied to select 136 probands (71 Stickler cases, 21 Spondyloepiphyseal dysplasia congenita cases, 11 Kniest dysplasia cases, and 34 other dysplasia cases) before molecular diagnosis by Sanger sequencing. We identified 66 different variants among the 71 positive patients. Among those patients, 18 belonged to multiplex families and 53 were sporadic. Most variants (38/44, 86%) were located in the triple helical domain of the collagen chain and glycine substitutions were mainly observed in severe phenotypes, whereas arginine to cysteine changes were more often encountered in moderate phenotypes. This series of skeletal dysplasia is one of the largest reported so far, adding 44 novel variants (15%) to published data. We have confirmed that about half of our Stickler patients (46%) carried a COL2A1 variant, and that the molecular spectrum was different across the phenotypes. To further address the question of genotype–phenotype correlation, we plan to screen our patients for other candidate genes using a targeted next-generation sequencing approach.
Similar content being viewed by others
Introduction
Collagen, type II, alpha 1 is the major collagen type synthesized by chondrocytes and the adult vitreous. Type II collagenopathies include a wide variety of skeletal dysplasia ranging from lethal disorders, such as achondrogenesis, type II or hypochondrogenesis (MIM#200610), to adult early onset osteoarthritis at the milder end of the spectrum. Stickler syndrome type I, STL1 (MIM#108300) and Stickler syndrome type I nonsyndromic predominantly ocular (MIM#609508) are the most frequent forms of type II collagenopathies. Stickler syndrome has an estimated incidence ranging from 1 in 7500 to 1 in 9000 births (http://ghr.nlm.nih.gov/condition/stickler-syndrome). Stickler syndrome is characterized by the association of spondyloepiphyseal dysplasia, joint laxity, myopia with congenital vitreous anomalies, and the risk of retinal detachment. These are in addition to conductive and sensorineural progressive hearing loss occurring as early as in the first year of life.
Molecular defects in the COL2A1 gene (MIM#108300), which encodes the alpha 1 chain of procollagen type II, result in skeletal, orofacial, and ocular disorders. These dominantly inherited diseases are associated with various COL2A1 variants that include missense, nonsense, small indels or large deletions.1, 2 The identification of the causative variant in the COL2A1 gene is essential for establishing the early management of at-risk individuals to propose an accurate assessment of the risks of recurrence in siblings and possibly a prenatal diagnosis.
In 2011, our laboratories developed the molecular screening of the COL2A1 gene and became the managers of a database specific for this gene at http://databases.lovd.nl/shared/genes/COL2A1, which belongs to the LOVD (Leiden Open Variation Database) project. Most of the related published studies are focused on variants identified in Stickler patients. In a recent series of 188 Stickler patients, more than half (N=100) were found to harbor a variant in the COL2A1 gene.1 However, large series of the other phenotypes associated with variants in this gene have never been reported. In the present study, we describe the molecular spectrum of a total of 136 probands referred to for type II collagenopathies.
Materials and methods
Clinical assessment
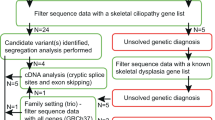
All patients referred to our laboratories (Laboratoire de génétique des maladies rares et auto-inflammatoires, CHRU, Montpellier, France) for suspected type II collagenopathy were assessed by a local expert clinician, Professor David Geneviève (DG), the Coordinator of the competence Centre of rare diseases for skeletal dysplasia for the South of France at Montpellier, before molecular screening of the COL2A1 gene. Demographic, clinical and radiographic data were collected from probands using a specific questionnaire available online at http://umai.chu-montpellier.fr/diag2.html. DNA or blood samples were requested from all 136 probands and relevant relatives if available (Figure 1). Informed consent was obtained from each participant before the genetic analysis.
Point variant screening and validation
Genomic DNA was isolated from peripheral EDTA anti-coagulated whole blood using QIAamp DNA Blood Kits (Qiagen, Valencia, CA, USA). Two sets of primer pairs (sequences available upon request) were designed for the COL2A1 gene (NG_008072.1) using the Primer-Blast software (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). Universal M13 tags were added at the 5′ ends of each primer. Polymerase chain reactions (PCRs) were performed with the Promega PCR Master Mix according to the manufacturer’s protocol (Promega, Madison, WI, USA). The entire coding sequence and intronic boundaries were screened for variants by direct sequencing of two PCR products using the BigDye Terminator v3.1 Cycle Sequencing Kit and M13 primers, followed by electrophoresis of the amplicons on an ABI 3100XL Genetic Analyzer according to the manufacturer’s recommendations (Applied Biosystems, Foster City, CA, USA). The obtained sequences were aligned against the COL2A1 gene reference sequences from NCBI (NG_008072.1 and NM_001844.4) using the SeqScape software.
When a putative disease-causing variant was identified, we performed parental analysis when possible to evaluate segregation and de novo occurrence, checked the variant frequency in healthy individuals from public databases (1000 Genomes, Exome Variant Server NHLBI GO Exome Sequencing Project, dbSNP), and the variant pathogenicity using in silico predictive tools (Polyphen-2: http://genetics.bwh.harvard.edu/pph2, SIFT: http://sift.bii.a-star.edu.sg, MutationTaster: http://www.mutationtaster.org/ and Human Splicing Finder: 'http://www.umd.be/HSF3/).
Rearrangement screening by MLPA and quantitative PCR confirmation
Multiplex Ligation-dependent Probe Amplification (MLPA) was performed according to the manufacturer’s recommendations (MRC Holland, Amsterdam, The Netherlands). When Sanger sequencing identified no point COL2A1 variant, patients’ DNA were analyzed using the probe set for COL2A1 (SALSA MLPA kit 214).
Deletions and duplications detected by MLPA were then confirmed with a qPCR assay using specific primers designed to produce amplicons ranging from 100 to 400 bp on a LightCycler 480 instrument (Roche diagnostics, Meylan, France), as previously described.3 To specify the extent of the whole gene deletion, we used a CytoScanHD array from Affymetrix, according to the supplier’s instructions.
Results
Phenotype distribution
Clinical charts from the patients referred for possible type II collagenopathy were reviewed and ascertained by our clinical referral (DG). Among them, 136 probands showed the clinical and radiological features of type II collagenopathies. Almost all the COL2A1-related skeletal disorders were represented and probands were classified as summarized in Table 1. About half of our patients had Stickler syndrome.
Variant distribution and type
We analyzed the entire coding sequence of the COL2A1 gene and its intronic boundaries, identifying at least one variant in 71 (52%) of our 136 probands (Figure 1). We observed variants in 8 of 9 (89%) Spondyloepimetaphyseal dysplasia Strudwick type (SEMDSTWK) cases, 15 of 21 (71%) Spondylo epiphyseal dysplasia congenita cases (SEDC), 6 of 9 (67%) of the achondrogenesis, type II or hypochondrogenesis cases, 7 of 11 (64%) of the Kniest dysplasia cases, 33 of 71 (46%) of the Stickler patients, and 2 of 5 (40%) of the early onset osteoarthritis (Figure 2a).
(a) Variant distribution according to the patients’ phenotype. Achondrogenesis, type II or hypochondrogenesis (MIM#200610), Czech dysplasia (MIM#609162), Kniest dysplasia (MIM#156550), SEMD Strudwick type (MIM#184250), SpondyloEpiphyseal Dysplasia Congenita (MIM#183900), Stickler type 1 (MIM#108300; 609508). (b) Variant distribution among patients according to the type and effect of the variant. (c) Distribution of the variant type and effect according to the patients’ phenotype.
The 71 positive probands displayed a total of 66 different variants (Table 1 and Figure 2b). Missense variants were the most frequently observed (58%), including 28 glycine substitutions within a Gly-X-Y repeat. Patients in whom we did not detect point variation by Sanger sequencing were further assessed for gene rearrangement that revealed, in three cases, deletions hereafter termed ‘large deletions’.
Novel variants
Among the 66 different variants, 44 were novel (Table 1) and distributed throughout the gene; 38 of 44 (86%) of these are mainly located in the triple helical domain of the procollagen α1 (II)-chain (Figure 3). These 44 novel variants included 26 missense variants and three large deletions. All but one p.(Ala340Thr), c.1018G>A, which was also identified in an unaffected parent) were predicted as likely to be deleterious using the PolyPhen-2 and Sift online programs (http://genetics.bwh.harvard.edu/pph2/ and http://sift.jcvi.org/). Interestingly, in a Stickler family, the p.(Pro314Argfs*315) variant carried by the proband was inherited from a mosaic father (Table 1).
Distribution of the 44 novel COL2A1 variants identified in 136 French patients with skeletal dysplasia. I, intron; E, exon; bp, base pair; aa, amino acid; bracket, large deletion; circle, premature stop codon; triangle, small rearrangement; square, glycine missense; diamond, nonglycine missense and asterisk, RNA processing.
Attempts to genotype/phenotype correlations
When we plotted the variant type against the patient’s phenotype, we noticed that the molecular spectrum was different across the diseases. All variant types were observed in Stickler syndrome, whereas only missense variants were observed in SEDC (Figure 2c).
Five arginine to cysteine substitutions were identified in phenotypes with various severities (Table 1). p.(Arg550Cys), c.1648C>T, and p.(Arg719Cys), c.2155C>T were found in two of the five early onset osteoarthritis cases, p.(Arg989Cys), c.2965C>T was found in one Strudwick case, p.(Arg904Cys), c.2710C>T was found in one Stickler case and one Kniest case, and p.(Arg137Cys), c.409C>T was found in a fetus with achondrogenesis, type II. Among the other novel missense variants, we identified three proline substitutions within the Gly-X-Y repeat: p.(Pro133His), c.398C>A was carried by a Stickler patient and her affected mother, p.(Pro434Ser), c.1300C>T was identified in a Kniest patient inherited from his asymptomatic father, and a homozygous variant p.(Pro458Leu), c.1373C>T was found in a SEDC case, inherited from both parents (Table 1).
Among the seven splice variants putatively affecting RNA processing, five were identified in Stickler patients and the two others were identified in a SEMD Strudwick patient and a Kniest patient.
Ten of 11 frameshift variants resulting from small deletions or duplications were identified in Stickler patients. One frameshift variant, p.(Ala895Serfs*49), c.2678dupC, was carried by a Kniest patient.
We also identified three large deletions in two of the most extreme phenotypes, namely a 1364-bp in-frame deletion spanning exons 42 to 46, p.(Pro916_Gly1074del), c.2745_3221del, in a patient with achondrogenesis, type II, a frameshift deletion spanning introns 1 to 16, p.(Gln29Argfs*35), c.85+1360_1023+555del, and a deletion of the entire gene, p.0, c.(?_-181)_(*442_?)del, in two patients with Stickler syndrome (Table 1).
Variant segregation analysis
Our series included 71 positive cases, among which 53 were sporadic and 18 were from multiplex families (with more than one affected subject; Figure 1).
Sporadic cases
DNA from both parents was available for 16 sporadic cases. The variant was found to be a priori de novo in 15 cases, but was detected in the asymptomatic mother of one achondrogenesis, type II fetus carrying p.(Arg137Cys), c.409C>T (Figure 1).
Familial cases
Among the 42 available relatives of the multiplex families, 15 were asymptomatic and 27 were symptomatic (Figure 1). Of those 27 symptomatic relatives, 26 carried the proband’s variant and 1 did not, whereas one of the 15 asymptomatic relatives displayed the proband’s variant p.(Ala340Thr), c.1018G>A. These observations support the genetic heterogeneity and the existence of a variant of uncertain significance, respectively. Two representative multiplex families are shown in Figure 4.
Pedigree of two representative Stickler multiplex families. (a) Segregation of the p.(Gly366Alafs*263) (c.1095delT) frameshift deletion. (b) Segregation of the intronic c.2355+4A>C variant. Open and dark symbols denote asymptomatic and affected subjects, respectively. The year of birth and the variant status are shown below the symbols.
Discussion
In this work, we reported the screening for mutants of the COL2A1 gene in a cohort of 136 probands with clinical and/or radiographic suspicion of a type II collagen disorder. A majority 71 of 136 (52%) of this large cohort were Stickler patients, but 20 of 136 (15%) of the cases were SEDC patients and 5 of 136 (4%) were early onset osteoarthritis patients (Table 1). We identified 66 different COL2A1 variants spread throughout the COL2A1 gene. Forty-four of these were novel, which substantially adds to an approximate 300 COL2A1 variants previously reported in the LOVD database (Table 1) and in the literature.1, 2
No variant hotspot was observed in our study, as previously reported.1 Glycine changes represented 19 of 44 (43%) of the novel variants and were all located in a triple helical region of the collagen 2 (Table 1, Figure 3). The substitution of glycine with bulkier amino acids in a Gly-X-Y repeat were more often associated with severe phenotypes, such as achondrogenesis, type II or hypochondrogenesis, or with severe short stature phenotypes including Kniest, SEDC and SEMD Strudwick types.4, 5, 6 Counter to previous reports, variants in the X or Y positions of the Gly-X-Y motif in the collagen chain did not seem to influence the phenotypes observed in our patients.
Three new proline substitutions were identified in different phenotypes, which were p.(Pro133His), c.398C>A, in Stickler, p.(Pro434Ser), c.1300C>T, in Kniest, and p.(Pro458Leu), c.1373C>T, in SEDC (Table 1). Although the former was located in the X position of the repeat found in the N-terminal region, the two other variants were in the Y position of the repeats found in the triple helical domain. Interestingly, another proline variant located in the Y position p.(Pro986Leu), c.2957C>T was previously shown to segregate with mild SEDC in a three-generation family.7 Proline is a highly conserved amino acid in the Y position that contributes to the thermal stability of the triple helix.8 Thus, the replacement of the proline with a leucine in the Y position might affect the collagen structure and likely accounts for the short stature observed in dysplasia. However, the pathogenicity of the Pro434Ser substitution remains uncertain, as it was inherited from an asymptomatic parent.
We also report five arginine to cysteine variants, including p.(Arg137Cys), c.409C>T; p.(Arg550Cys), c.1648C>T; p.(Arg719Cys), c.2155C>T; p.(Arg904Cys), c.2710C>T; and p.(Arg989Cys), c.2965C>T (Table 1). The first one, p.(Arg137Cys), c.409C>T, is novel and was identified in a fetus with achondrogenesis, type II. Although it was predicted to be pathogenic by the Polyphen program or damaging by the Sift program, this variant was inherited from the healthy mother. Thus, we propose that it either does not cause the disease phenotype or is not fully penetrant. In accordance with this hypothesis, it has been reported that unlike the other four variants, p.(Arg137Cys), c.409C>T is not located within the critical triple helical region of the collagen chain, and arginine to cysteine variants never result in lethal conditions.9 Sequencing the DNA of this fetus for DTDST/SLC26A2, a gene associated with achondrogenesis, type Ib, was also negative. We are currently developing next-generation sequencing using a custom panel of 29 candidate genes that include GDF5 and FGFR3 to resolve this case and in the cases in which no COL2A1 variant was identified. Two other arginine to cystein variants, p.(Arg550Cys), c.1648C>T and p.(Arg719Cys), c.2155C>T, were observed in two early onset osteoarthritis probands and both segregated with the disease (manuscript in preparation). p.(Arg550Cys), c.1648C>T was novel, but p.(Arg719Cys), c.2155C>T has already been reported once in a patient with osteoarthritis and mild chondrodysplasia.10 Eyre et al.11 have demonstrated that p.(Arg719Cys), c.2155C>T is responsible for a reduction in protein secretion and a limited collagen assembly. The fourth arginine to cystein variant, p.(Arg904Cys), c.2710C>T, was found once in a Stickler case and once in a Kniest case, from our series. This variant was already observed in several Stickler cases.9, 12, 13, 14 Finally, p.(Arg989Cys), c.2965C>T was carried by one of our SEMD Strudwick patients, and this variant has been previously reported in several SEDC cases.9, 15, 16, 17 In accordance with the literature, we observed that arginine to cysteine changes were associated with a broad spectrum of diseases, but they were more often encountered in the moderate phenotypes.9 Nevertheless, our data confirm that variants such as p.(Arg904Cys), c.2710C>T and p.(Arg989Cys), c.2965C>T, located near the carboxyl terminus of the collagen triple helix were associated with more severe phenotypes.9 Arginine to cysteine substitutions introduce novel cysteine residues into the triple helical region, leading to aberrant disulfide bonds between mutant pro-collagen chains.1 It has also been shown that arginine changes located within a thermostable region such as p.(Arg989Cys), c.2965C>T alter both the assembly of collagen into fibrils and their interaction with other proteins of the cartilage matrix.18 Hence, these variants could disturb the thermostability of the protein and exert a dominant negative effect.
We have also identified 11 small deletions and duplications that result in frameshift variants and premature stop codons (PTC; Figure 2c). The p.(Ala895Serfs*49), c.2678dupC variant was carried by one of our Kniest patients, but this variant was previously identified in a Stickler case.1 The p.(Pro314Argfs*315), c.941delC variant was transmitted from a mosaic Stickler father to his affected young daughter who, unlike her father, presented with myopia. One-third (10/33) of our Stickler patients carried PTC variants. Such variants are usually subject to nonsense-mediated decay (NMD), resulting in a loss of function. This mechanism of type II collagen deficiency had often been reported in cases of milder skeletal dysplasia such as Stickler syndrome.1 In addition to small rearrangements, we also identified three large deletions (Figures 2b and c). The first was a whole gene deletion detected in a patient with a complex phenotype that included Stickler features. This deletion encompassed a wide region of 2.23 Mb on the chromosome 12q13.11 (GRCh37/hg19, chr12:46,316,398..48,547,316), which contained several genes around the COL2A1 gene (data not shown). Deletions of the entire COL2A1 gene and its boundaries have been previously published.19, 20, 21, 22 The second large (10.554 bp) deletion, p.(Gln29Argfs*35), c.85+1360_1023+555del, was identified in a Stickler proband and his affected father. Both patients displayed severe myopia, and the proband also had bilateral retinal detachment and skeletal anomalies. Thus, the phenotypic differences between the proband and his father suggest an intra-familial variance in clinical expression, suggesting the involvement of modifiers.23 These two deletions, which were both identified in Stickler cases, were also predicted to be subjected to NMD like the smaller rearrangement described above and would result in a haploinsufficiency.19 The third large deletion was an in-frame deletion p.(Pro916_Gly1074del), c.2745_3221del, that was expected to result in a shortened peptide lacking 159 amino acids that would exert a dominant negative effect on the ‘healthy’ component of the trimeric collagen. This hypothesis might account for the severe phenotype observed in this lethal achondrogenesis, type II case.
Splice site variants potentially lead to an abnormal RNA processing that could result in out-of-frame insertions or deletions, creating PTC subjected to NMD. The underlying pathogenic mechanism is therefore a loss of function often associated with mild phenotypes such as Stickler syndrome.1 Consistently, five of the seven RNA-processing variants we reported (Table 1) were all identified in Stickler cases. These were c.1366-2A>T, c.2049+1G>A, c.2193+2T>C, c.2355+4A>C, and c.4074+2_4074+3delTG. Moreover, c.2355+4A>C segregated in one multiplex Stickler family (Figure 4). The sixth c.1365+1G>C splice variant was carried by a Kniest dysplasia patient. Interestingly, the seventh variant c.709-2A>G was carried by a SEMD Strudwick patient. This latter variant was previously reported in an oto-spondylo-megaepiphyseal dyplasia case (OSMED, #215150) and was shown to cause exon skipping, resulting in short transcripts that escaped NMD and led to an in-frame deletion of the triple helical region of the collagen chain.24
Our observations provide a nearly 15% increase in the number of novel variants shown in previous records and illustrate the wide mutability of the COL2A1 gene and its various associated phenotypes.25 Our results also demonstrate the limits of focusing on a single gene during genetic diagnosis given the lack of clear phenotype to genotype correlations. To address this issue, we are now implementing next-generation sequencing approaches in our routine settings.24
References
Hoornaert KP, Vereecke I, Dewinter C et al: Stickler syndrome caused by COL2A1 mutations: genotype-phenotype correlation in a series of 100 patients. Eur J Hum Genet EJHG 2010; 18: 872–880.
Richards AJ, Laidlaw M, Whittaker J et al: High efficiency of mutation detection in type 1 stickler syndrome using a two-stage approach: vitreoretinal assessment coupled with exon sequencing for screening COL2A1. Hum Mutat 2006; 27: 696–704.
Pallares-Ruiz N, Philibert L, Dumont B et al: Combined mutation and rearrangement screening by quantitative PCR high-resolution melting: is it relevant for hereditary recurrent Fever genes? PloS One 2010; 5: e14096.
Mortier GR, Weis M, Nuytinck L et al: Report of five novel and one recurrent COL2A1 mutations with analysis of genotype-phenotype correlation in patients with a lethal type II collagen disorder. J Med Genet 2000; 37: 263–271.
Tysoe C, Saunders J, White L et al: A glycine to aspartic acid substitution of COL2A1 in a family with the Strudwick variant of spondyloepimetaphyseal dysplasia. QJM 2003; 96: 663–671.
Nishimura G, Haga N, Kitoh H et al: The phenotypic spectrum of COL2A1 mutations. Hum Mutat 2005; 26: 36–43.
Mark PR, Torres-Martinez W, Lachman RS, Weaver DD : Association of a p.Pro786Leu variant in COL2A1 with mild spondyloepiphyseal dysplasia congenita in a three-generation family. Am J Med Genet A 2011; 155A: 174–179.
Berg RA, Prockop DJ : The thermal transition of a non-hydroxylated form of collagen. Evidence for a role for hydroxyproline in stabilizing the triple-helix of collagen. Biochem Biophys Res Commun 1973; 52: 115–120.
Hoornaert KP, Dewinter C, Vereecke I et al: The phenotypic spectrum in patients with arginine to cysteine mutations in the COL2A1 gene. J Med Genet 2006; 43: 406–413.
Ala-Kokko L, Baldwin CT, Moskowitz RW, Prockop DJ : Single base mutation in the type II procollagen gene (COL2A1) as a cause of primary osteoarthritis associated with a mild chondrodysplasia. Proc Natl Acad Sci USA 1990; 87: 6565–6568.
Eyre DR, Weis MA, Moskowitz RW : Cartilage expression of a type II collagen mutation in an inherited form of osteoarthritis associated with a mild chondrodysplasia. J Clin Invest 1991; 87: 357–361.
Ballo R, Beighton PH, Ramesar RS : Stickler-like syndrome due to a dominant negative mutation in the COL2A1 gene. Am J Med Genet 1998; 80: 6–11.
Richards AJ, McNinch A, Martin H et al: Stickler syndrome and the vitreous phenotype: mutations in COL2A1 and COL11A1. Hum Mutat 2010; 31: E1461–E1471.
Nagendran S, Richards AJ, McNinch A, Sandford RN, Snead MP : Somatic mosaicism and the phenotypic expression of COL2A1 mutations. Am J Med Genet A 2012; 158A: 1204–1207.
Chan D, Taylor TK, Cole WG : Characterization of an arginine 789 to cysteine substitution in alpha 1 (II) collagen chains of a patient with spondyloepiphyseal dysplasia. J Biol Chem 1993; 268: 15238–15245.
Donahue LR, Chang B, Mohan S et al: A missense mutation in the mouse Col2a1 gene causes spondyloepiphyseal dysplasia congenita, hearing loss, and retinoschisis. J Bone Miner Res Off J Am Soc Bone Miner Res 2003; 18: 1612–1621.
Zhang Z, He J-W, Fu W-Z, Zhang C-Q, Zhang Z-L : Identification of three novel mutations in the COL2A1 gene in four unrelated Chinese families with spondyloepiphyseal dysplasia congenita. Biochem Biophys Res Commun 2011; 413: 504–508.
Steplewski A, Ito H, Rucker E et al: Position of single amino acid substitutions in the collagen triple helix determines their effect on structure of collagen fibrils. J Struct Biol 2004; 148: 326–337.
Van Der Hout AH, Verlind E, Beemer FA, Buys CHCM, Hofstra RMW, Scheffer H : Occurrence of deletion of a COL2A1 allele as the mutation in Stickler syndrome shows that a collagen type II dosage effect underlies this syndrome. Hum Mutat 2002; 20: 236.
Miyake N, Tonoki H, Gallego M et al: Phenotype-genotype correlation in two patients with 12q proximal deletion. J Hum Genet 2004; 49: 282–284.
Yamanishi T, Nishio J, Miya S et al: 12q interstitial deletion with bilateral cleft lip and palate: case report and literature review. Cleft Palate Craniofac J 2008; 45: 325–328.
Gimelli S, Makrythanasis P, Stouder C, Antonarakis SE, Bottani A, Béna F : A de novo 12q13.11 microdeletion in a patient with severe mental retardation, cleft palate, and high myopia. Eur J Med Genet 2011; 54: 94–96.
Liberfarb RM, Levy HP, Rose PS et al: The Stickler syndrome: genotype/phenotype correlation in 10 families with Stickler syndrome resulting from seven mutations in the type II collagen gene locus COL2A1. Genet Med 2003; 5: 21–27.
Miyamoto Y, Nakashima E, Hiraoka H, Ohashi H, Ikegawa S : A type II collagen mutation also results in oto-spondylo-megaepiphyseal dysplasia. Hum Genet 2005; 118: 175–178.
Terhal PA, Nievelstein RJ, Verver EJ et al: A study of the clinical and radiological features in a cohort of 93 patients with a COL2A1 mutation causing spondyloepiphyseal dysplasia congenital or a related phenotype. Am J Med Genet A 2015; 167A: 461–475.
Kannu P, Irving M, Aftimos S, Savarirayan R : Two novel COL2A1 mutations associated with a Legg-Calvé-Perthes disease-like presentation. Clin Orthop Relat Res 2011; 469: 1785–1790.
Meredith SP, Richards AJ, Bearcroft P et al: Significant ocular findings are a feature of heritable bone dysplasias resulting from defects in type II collagen. Br J Ophthalmol 2007; 91: 1148–1151.
Richards AJ, Martin S, Yates JR et al: COL2A1 exon 2 mutations: relevance to the Stickler and Wagner syndromes. Br J Ophthalmol 2000; 84: 364–371.
Ahmad NN, Ala-Kokko L, Knowlton RG et al: Stop codon in the procollagen II gene (COL2A1) in a family with the Stickler syndrome (arthro-ophthalmopathy). Proc Natl Acad Sci USA 1991; 88: 6624–6627.
Hintze V, Steplewski A, Ito H, Jensen DA, Rodeck U, Fertala A : Cells expressing partially unfolded R789C/p.R989C type II procollagen mutant associated with spondyloepiphyseal dysplasia undergo apoptosis. Hum Mutat 2008; 29: 841–851.
Kuivaniemi H, Tromp G, Prockop DJ : Mutations in fibrillar collagens (types I, II, III, and XI), fibril-associated collagen (type IX), and network-forming collagen (type X) cause a spectrum of diseases of bone, cartilage, and blood vessels. Hum Mutat 1997; 9: 300–315.
Chan D, Cole WG : Low basal transcription of genes for tissue-specific collagens by fibroblasts and lymphoblastoid cells. Application to the characterization of a glycine 997 to serine substitution in alpha 1(II) collagen chains of a patient with spondyloepiphyseal dysplas. J Biol Chem 1991; 266: 12487–12494.
Zechi-Ceide RM, Jesus Oliveira NA, Guion-Almeida ML, Antunes LF, Richieri-Costa A, Passos-Bueno MR : Clinical evaluation and COL2A1 gene analysis in 21 Brazilian families with Stickler syndrome: identification of novel mutations, further genotype/phenotype correlation, and its implications for the diagnosis. Eur J Med Genet 2008; 51: 183–196.
Unger S, Korkko J, Krakow D et al: Double heterozygosity for pseudoachondroplasia and spondyloepiphyseal dysplasia congenita. Am J Med Genet 2001; 104: 140–146.
Acknowledgements
This work was supported by the CHRU de Montpellier. We thank the probands and their relatives for agreeing to participate in the molecular analysis and American Journal Experts for editing the manuscript for English language.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Barat-Houari, M., Dumont, B., Fabre, A. et al. The expanding spectrum of COL2A1 gene variants IN 136 patients with a skeletal dysplasia phenotype. Eur J Hum Genet 24, 992–1000 (2016). https://doi.org/10.1038/ejhg.2015.250
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2015.250
This article is cited by
-
Novel COL2A1 variants in Japanese patients with spondyloepiphyseal dysplasia congenita
Human Genome Variation (2022)
-
Expanding the clinical spectrum of COL2A1 related disorders by a mass like phenotype
Scientific Reports (2022)
-
A novel homozygous variant of COL2A1 in a Chinese male with type II collagenopathy: a case report
BMC Medical Genomics (2021)
-
Best practice guidelines in managing the craniofacial aspects of skeletal dysplasia
Orphanet Journal of Rare Diseases (2021)
-
Report on three additional patients and genotype–phenotype correlation in SLC25A22-related disorders group
European Journal of Human Genetics (2019)