Abstract
Continued ability to digest lactose after weaning provides a possible selective advantage to individuals who have access to milk as a food source. The lactase persistence (LP) phenotype exists at varying frequencies in different populations and SNPs that modulate the regulation of the LCT gene have been identified in many of these populations. Very strong positive selection for LP has been illustrated for a single SNP (rs4988235) in northwestern European populations, which has become a textbook example of the effect of recent selective sweeps on genetic variation and linkage disequilibrium. In this study, we employed two different methods to detect signatures of positive selection in an East African pastoralist population in the HapMap collection, the Maasai from Kenya, and compared results with other HapMap populations. We found that signatures of recent selection coinciding with the LCT gene are the strongest across the genome in the Maasai population. Furthermore, the genome-wide signal of recent positive selection on haplotypic variation and population differentiation around the LCT gene is greater in the Maasai than in the CEU population (northwestern European descent), possibly due to stronger selection pressure, but it could also be an indication of more recent selection in Maasai compared with the Central European group or more efficient selection in the Maasai due to less genetic drift for their larger effective population size. This signal of recent selection is driven by a putative East African LP haplotype that is different from the haplotype that contributes to the LP phenotype in northwestern Europe.
Similar content being viewed by others
Introduction
The continued ability to digest lactose after weaning varies among humans and it is particularly common among populations that have traditionally practiced cattle herding. The capacity of continued lactose digestion or lactase persistence (LP) is conferred to humans by a few mutations in a cis-acting control element of the LCT gene that encodes the lactase-phlorizin hydrolase enzyme (LPH).1, 2, 3 It has been shown that particular variants at SNPs in the introns of an adjacent gene (MCM6) prevent the downregulation of LPH in adults.4, 5, 6, 7 One of these SNPs (13910C-T or rs4988235) has likely been under strong selection in some European populations.8, 9, 10 Especially Northern Europeans show high frequencies of this mutation and simultaneously show high levels of LP. The frequency of this SNP-variant and the ability for adults to digest lactose decreases towards southern Europe and the Middle East and is low in North Africa.3, 6, 11 This particular mutation is at very low frequency or absent in sub-Saharan African populations even though some groups, such as East African pastoralists, show a high prevalence of LP.12
Subsequent candidate gene studies showed that a different polymorphism (14010G-C), 100 bp downstream from the SNP-variant that causes LP in Northern Europeans, which occurred on a different haplotypic background, was strongly linked to LP in various East African groups and that there was a strong signal for selection in some of these populations.7 The frequency of this variant varies between different East African groups and occurs at frequencies of 39 and 32% in Nilo-Saharans from Tanzania and Kenya and at frequencies of 46 and 18% in Afro-Asiatic groups from these two countries. The frequencies are lower in the Sandawe (13%) and absent in the Hadza hunter-gatherers from East Africa, as well as in various Sudanese populations.7 However, patterns of genome-wide genetic variation and linkage disequilibrium in East African populations remain poorly studied, and the signature of recent selection seen around the LCT locus in Nilo-Saharans and Afro-Asiatic groups has not yet been compared with other parts of the genome in these populations. West African farmers (such as the Yoruba from Nigeria) central and southern African hunter-gatherers, and East African Bantu-speaking groups show no signature of selection at the LCT locus.10, 13, 14 Both of these LP polymorphisms (European −13910C-T and East African −14010G-C) have been directly attributed to the enhancement of transcription of the LCT gene, by means of binding affinity and reporter gene assays.15, 16, 17 In addition to the two polymorphisms mentioned above, three other polymorphisms within the adjacent MCM6 gene have also been linked to the lactase persistent trait in specific groups of people. The compound −13712C, −13915G allele has a role in LP in the Middle East,4 while the −22018G-A SNP is linked to the trait in certain northern European populations5 and the −13907C-G SNP is linked to the trait in some Sudanese populations.7 Although the function of MCM6 is unrelated to the LCT gene function and the LP trait, it contains two of the regulatory regions for LCT, located in two of the MCM6 introns, ∼14 kb (most of the LP polymorphisms) and 22 kb (the −22018G-A variant) upstream of the LCT gene (a summary can be found at http://omim.org/entry/601806).
In this study, we performed genome-wide scans for recent positive selection in the HapMap Maasai population,18 including the region on chromosome 2q21 where the LCT and MCM6 genes are located and compared results with other HapMap populations. We found that signatures of recent selection at the LCT/MCM6 gene-region are the strongest across the genome in the Maasai population. Furthermore, the signals of recent positive selection around the LCT gene are stronger in the Maasai than in the CEU population, which can be caused by stronger selection pressure in the Maasai, more recent selection in the Maasai, or different demographic history of the Maasai and the CEU.
Materials and methods
We obtained phased genotype data comprising 1 387 465 autosomal SNPs from HapMap III18 for 204 individuals from 7 HapMap populations: CEU, TSI, MKK, LWK, YRI, JPT, and CHB (Downloaded 30 Nov, 2010: ftp://ftp.ncbi.nlm.nih.gov/hapmap/phasing/2009-02_phaseIII/HapMap3_r2/). We used Chimpanzee alleles from panTro219 in an alignment with the human genome20 to determine the ancestral SNP-variant. A genetic map was also retrieved from the 1000 genomes data (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/working/20110217_M broad_omni_genotypes/). We retained a final set of 466 614 SNPs that had both a genetic map position and the ancestral SNP-variant inferred from the Chimpanzee genome.
Integrated haplotype statistic (iHS) values,13 were calculated with the software iHS (http://hgdp.uchicago.edu/Software/) for the MKK, CEU, TSI, LWK and YRI groups. For a window-based measure ‘winiHS’, we calculated the mean of the absolute value of iHS in a window of 30 consecutive SNPs and with a 15 SNP jump length between windows.
The population branch statistic (PBS) was computed according to Yi et al.21 For each branch in the unrooted population topology connecting MKK, CEU and HapMap East Asians (JPT+CHB), the PBS statistic was computed using an allele frequency based estimate of FST.22 The average PBS in windows of 30 SNPs and step-length 15 SNPs was calculated based on the same SNPs as in the iHS-analysis, and standardized by subtracting the mean and dividing by the SD (of the window values) resulting in a statistic that we refer to as ‘window-based measure of PBS’ (winPBS).
A Median Joining Network (with Maximum Parsimony post-processing) was constructed using Network v.4.6.0.023, 24 for a ∼100 kb region encompassing the MCM6 gene, which contained 60 SNPs in the HapMap III data.
Results
We scanned the genome of five HapMap populations (MKK – Maasai, Nilo-Saharan speakers from Kenya in East Africa (n=87); CEU – western European ancestry (n=17); TSI – Tuscan from Italy (n=88); YRI – Yoruba, Niger-Kordofanian speakers from Nigeria in West Africa (n=9), and LWK – Luhya, Bantu-speakers from Kenya in East Africa (n=90)), for regions of extended haplotype homozygosity using iHS,13 which can detect selective sweeps that have not yet reached fixation in a population. Subsequently, we calculated a window-based statistic (winiHS) as explained in the ‘Materials and Methods’ section. The choice of window size had little impact on the result, see Supplementary Figure S1.
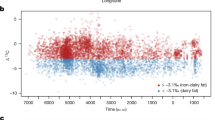
Across the entire genome for the MKK, the strongest winiHS signal was found in the chromosomal region where the LCT gene is located (Figure 1a). Furthermore, the top 16 strongest genome-wide winiHS signals were confined to this region. The signal in this region was absent in all populations except for MKK and CEU (Figure 1b, Supplementary Figure S2, Supplementary Table S1). The strongest signal in the TSI, the YRI and the LWK was found at the MHC region (at ∼30 Mb on chromosome 6, 6p22.1), but the MKK and the CEU also showed strong signals in this region (Supplementary Figure S2, Supplementary Table S1). The top 20 winiHS peaks in all screened populations are shown in Supplementary Table S1. The signal in the LCT/MCM6 region has only a slightly longer extension in MKK (4.16 Mb) than CEU (3.3 Mb) but the peak winiHS value in MKK was about twice as high as in CEU (Figure 1b). While sample size differences between populations are a potential concern, the signal in MKK remained stronger than in CEU when down-sampling MKK to 34 haploid genomes (the same as for CEU) (Supplementary Figure S3).
Selection scans. (a) winiHS across the genome (see text) of the MKK population. Light blue corresponds to odd-numbered chromosomes while even-numbered chromosomes are colored dark blue. The LCT/MCM6 region on chromosome 2 and the MHC regions on chromosome 6 are marked by horizontal lines. (b) Close-up of winiHS in the lactase region (position 125 Mb to 145 Mb on chromosome 2). The numbers correspond to the rank across the entire genome of each population of the winiHS for the SNPs (the top 20 SNPs are shown). The gray vertical line marks the region of LCT/MCM6. (c) Close-up of the winPBS in the lactase region (position 125 Mb to 145 Mb on chromosome 2). The numbers correspond to the rank across the entire genome (the top 10 SNPs are shown).
To study the possible impact of recent positive selection in the East African and European populations using an alternative approach, we employed a method based on searching for unusually differentiated genomic regions using the PBS.21 A winPBS-value was computed as explained in the Methods section. This statistic revealed an unusually high differentiation of the MKK and CEU around the LCT region (Figure 1c), for which the peak winPBS values were greater in the MKK than in the CEU. The winPBS value in this region was the third strongest across the entire genome in the MKK sample.
Subsequently we focused on a ∼100 kb region encompassing the MCM6 gene, which contained 60 SNPs in the HapMap 3 data. A direct comparison of the haplotypes in the CEU subset and the MKK subset indicated that both these populations contained one specific high frequency haplotype, and that these two haplotypes differed substantially from each other. To visualize related haplotypes, we constructed a Median Joining Network23, 24 (Figure 2). The European LP variant (13910C-T at rs4988235) coincided with the most frequent CEU haplotype. The SNP associated with LP in East Africa7 was not present in our filtered data set nor in the complete HapMap3 data, but we identified a haplotype that putatively contain the East African LP causing variant. This haplotype is the most common haplotype (65.5%) in the Maasai (the second most common haplotype had a frequency of 7.5% and frequencies of the remaining 27 haplotypes were all below 3%). Owing to the exceptionally strong signal for selection that we observe, it is unlikely that any of these lower frequency haplotypes underlies the LP trait in the Maasai group. Furthermore, Tishkoff et al7 found the LP trait to be at frequencies of 71% and 59% in the Kenyan and Tanzanian Maasai, respectively (the frequency of the suggested HapMap Maasai LP haplotype is intermediate to these frequencies), and identified the −14010G-C mutation in 58% and 44.7% of the two respective groups. Finally, the genome of one of the HapMap Maasai individuals carrying the putative Maasai LP haplotype (NA21733), has been sequenced by Complete Genomics (http://www.completegenomics.com/). For the 100 kb region encompassing the MCM6 gene, this individual is homozygous for the most frequent Maasai haplotype (based on the HapMap data) and at the East African LP SNP, the individual carried one copy of the LP variant (−14010C) and one copy of the non-LP variant (−14010G). For these reasons, it is likely that the East African −14010C LP causing variant occurs (very often) on this high frequency haplotype-background in the Maasai. The other four identified LP SNPs were either absent or at very low frequencies in the two Maasai groups studied by Tishkoff et al.7
Haplotype network and frequency table for 60-SNP haplotypes encompassing the LCT and MCM6 loci. The network shows the relationship of the 60-SNP haplotypes in five selected HapMap3 populations. The CEU LP haplotype is indicated in the figure, as defined by the 13910C-T mutation (rs4988235). The inset-table shows the frequencies of the CEU LP haplotype, the MKK LP haplotype and other haplotypes in five different HapMap populations. The average PBS for the 60 SNPs had a greater value for MKK (0.57) than for CEU (0.43), where the genome average PBS values were 0.071 for MKK and 0.036 for CEU.
The East African haplotype putatively associated with LP in the Maasai, also occurred at high frequency (31.3%) in the HapMap Tuscans (TSI), three times as common as the northwestern European LP haplotype (10.2%, Figure 2).
Discussion
The LP phenotype confers a great advantage to individuals that live in pastoralist societies as it allows access to a new sustenance niche that would have otherwise been inaccessible. In addition, milk as a food source is more sustainable than meat production, with no need to cull in order to access the food. The HapMap Maasai population from Kenya is an East African pastoralist population that relies heavily on milk consumption as a food source, in addition to meat and blood.25, 26 Although meat is considered an important food source among the Maasai, it is consumed infrequently as personal wealth is measured in terms of cattle. In such a subsistence- and cultural background it is expected that the acquisition of LP will be highly advantageous.
In this study, we found the strongest genome-wide signal for selection at the LCT/MCM6 region in the HapMap Maasai using iHS selection scans. The only other HapMap population that showed a signal for selection in this region was the CEU group. Two different statistics that detect selection, iHS and PBS, indicated a stronger signal in the East African Maasai group compared with the European CEU group. The strong iHS signal might indicate stronger selection pressure in the Maasai, but it can also be an indication of more recent selection in Maasai compared with the CEU group or more efficient selection in the Maasai due to less genetic drift (larger Ne in Maasai). Indeed, Tishkoff et al,7 estimated a younger date (∼2700–6800) for the East African 14010C allele compared the European 13910T allele (∼8000–9000 years), although the time estimates had large overlapping confidence intervals. Furthermore, other factors such as different demographic histories of the two groups (ie, differences in effective population sizes and migration rates from neighboring populations) and the influence of ascertainment bias might also have a role, but the signal of selection is nevertheless stronger in the Maasai than in the CEU.
The haplotype network illustrated the two different haplotype backgrounds for the European and putative East African LP causing variants as was found by Tishkoff et al.7 The putative East African Maasai LP haplotype is at lower frequencies in the two other African groups (Yoruba and Luhya) and only one Northern European CEU individual carried this haplotype (Figure 2). The Tuscan group showed a higher frequency (31.3%) for this haplotype, though the iHS scan of the TSI did not show any signal of selection at the LCT locus (Supplementary Figure S1). However, LP has been shown to be present in 39.5% of Italians8, 11, 27 and the CEU LP haplotype was only present at 10.2% in Tuscans, which suggest that other polymorphisms might also be involved in the LP phenotype for this population. While the frequency of the putative East African LP haplotype is high in the Tuscans, without a direct survey of the −14010C LP causing variant in Tuscans, we can only speculate about the potential LP causing variant(s) in that population.
To conclude, our study documents a strong impact of recent positive selection on haplotype structure, variation, and differentiation associated with LP in the East African Maasai, and the genome-wide selection signal is greater than for the well-studied case of LP in Northwestern Europe.
References
Ingram CJ, Mulcare CA, Itan Y, Thomas MG, Swallow DM : Lactose digestion and the evolutionary genetics of lactase persistence. Hum Genet 2009; 124: 579–591.
Ingram CJ, Raga TO, Tarekegn A et al. Multiple rare variants as a cause of a common phenotype: several different lactase persistence associated alleles in a single ethnic group. J Mol Evol 2009; 69: 579–588.
Swallow DM : Genetics of lactase persistence and lactose intolerance. Annu Rev Genet 2003; 37: 197–219.
Enattah NS, Jensen TG, Nielsen M et al. Independent introduction of two lactase-persistence alleles into human populations reflects different history of adaptation to milk culture. Am J Hum Genet 2008; 82: 57–72.
Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Jarvela I : Identification of a variant associated with adult-type hypolactasia. Nat Genet 2002; 30: 233–237.
Enattah NS, Trudeau A, Pimenoff V et al. Evidence of still-ongoing convergence evolution of the lactase persistence T-13910 alleles in humans. Am J Hum Genet 2007; 81: 615–625.
Tishkoff SA, Reed FA, Ranciaro A et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet 2007; 39: 31–40.
Bersaglieri T, Sabeti PC, Patterson N et al. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet 2004; 74: 1111–1120.
Poulter M, Hollox E, Harvey CB et al. The causal element for the lactase persistence/non-persistence polymorphism is located in a 1 Mb region of linkage disequilibrium in Europeans. Ann Hum Genet 2003; 67: 298–311.
The International Haplotype Map Project A haplotype map of the human genome. Nature 2005; 437: 1299–1320.
Itan Y, Jones BL, Ingram CJ, Swallow DM, Thomas MG : A worldwide correlation of lactase persistence phenotype and genotypes. BMC Evol Biol 2010; 10: 36.
Mulcare CA, Weale ME, Jones AL et al. The T allele of a single-nucleotide polymorphism 13.9 kb upstream of the lactase gene (LCT) (C-13.9kbT) does not predict or cause the lactase-persistence phenotype in Africans. Am J Hum Genet 2004; 74: 1102–1110.
Voight BF, Kudaravalli S, Wen X, Pritchard JK : A map of recent positive selection in the human genome. PLoS Biol 2006; 4: e72.
Jakobsson M, Scholz SW, Scheet P et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature 2008; 451: 998–1003.
Jensen TG, Liebert A, Lewinsky R, Swallow DM, Olsen J, Troelsen JT : The −14010*C variant associated with lactase persistence is located between an Oct-1 and HNF1alpha binding site and increases lactase promoter activity. Hum Genet 2011; 130: 483–493.
Lewinsky RH, Jensen TG, Moller J, Stensballe A, Olsen J, Troelsen JT : T-13910 DNA variant associated with lactase persistence interacts with Oct-1 and stimulates lactase promoter activity in vitro. Hum Mol Genet 2005; 14: 3945–3953.
Olds LC, Sibley E : Lactase persistence DNA variant enhances lactase promoter activity in vitro: functional role as a cis regulatory element. Hum Mol Genet 2003; 12: 2333–2340.
Altshuler DM, Gibbs RA, Peltonen L et al. Integrating common and rare genetic variation in diverse human populations. Nature 2010; 467: 52–58.
The Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 2005; 437: 69–87.
Schuster SC, Miller W, Ratan A et al. Complete Khoisan and Bantu genomes from southern Africa. Nature 2010; 463: 943–947.
Yi X, Liang Y, Huerta-Sanchez E et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science 2010; 329: 75–78.
Weir BS, Cockerham CC : Estimating F-statistics for the analysis of population structure. Evolution 1984; 38: 1358–1370.
Bandelt HJ, Forster P, Rohl A : Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 1999; 16: 37–48.
Polzin T, Daneschmand SV : On Steiner trees and minimum spanning trees in hypergraphs. Operations Res Lett 2003; 31: 12–20.
Agriculture and Consumer ProtectionNutritional importance of traditional milk products in the national diet. The technology of traditional milk products in developing countries: http://www.fao.org/docrep/003/t0251e/T0251E07.htmFood and Agricultural Organization of the United Nations.
Kerven C : Some research and development implications for pastoral dairy production in Africa. ILCA Bulletin 1987; 26: 29–35.
Anagnostou P, Battaggia C, Coia V et al. Tracing the distribution and evolution of lactase persistence in Southern Europe through the study of the T(-13910) variant. Am J Hum Biol 2009; 21: 217–219.
Acknowledgements
We thank Agnes Sjöstrand and Lucie Gattepaille for helpful discussions. The computations were performed on resources provided by SNIC through Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) under Project p2011187. We thank the Swedish Research Council, the Swedish Research Council FORMAS, and the Wenner-Gren foundations for financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Schlebusch, C., Sjödin, P., Skoglund, P. et al. Stronger signal of recent selection for lactase persistence in Maasai than in Europeans. Eur J Hum Genet 21, 550–553 (2013). https://doi.org/10.1038/ejhg.2012.199
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2012.199
Keywords
This article is cited by
-
Spatial and temporal diversity of positive selection on shared haplotypes at the PSCA locus among worldwide human populations
Heredity (2023)
-
Male-biased migration from East Africa introduced pastoralism into southern Africa
BMC Biology (2021)
-
No Evidence of Copy Number Variation in Acidic Mammalian Chitinase Genes (CHIA) in New World and Old World Monkeys
International Journal of Primatology (2018)
-
World-wide distributions of lactase persistence alleles and the complex effects of recombination and selection
Human Genetics (2017)
-
Validating niche-construction theory through path analysis
Archaeological and Anthropological Sciences (2016)