Abstract
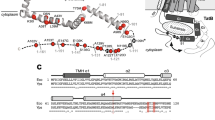
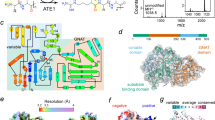
The twin-arginine translocation (Tat) pathway is one of two general protein transport systems found in the prokaryotic cytoplasmic membrane and is conserved in the thylakoid membrane of plant chloroplasts. The defining, and highly unusual, property of the Tat pathway is that it transports folded proteins, a task that must be achieved without allowing appreciable ion leakage across the membrane. The integral membrane TatC protein is the central component of the Tat pathway. TatC captures substrate proteins by binding their signal peptides. TatC then recruits TatA family proteins to form the active translocation complex. Here we report the crystal structure of TatC from the hyperthermophilic bacterium Aquifex aeolicus. This structure provides a molecular description of the core of the Tat translocation system and a framework for understanding the unique Tat transport mechanism.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Park, E. & Rapoport, T. A. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 41, 21–40 (2012)
Palmer, T. & Berks, B. C. The twin-arginine translocation (Tat) protein export pathway. Nature Rev. Microbiol. 10, 483–496 (2012)
Frobel, J., Rose, P. & Muller, M. Twin-arginine-dependent translocation of folded proteins. Phil. Trans. R. Soc. Lond. B 367, 1029–1046 (2012)
Celedon, J. M. & Cline, K. Intra-plastid protein trafficking; how plant cells adapted prokaryotic mechanisms to the eukaryotic condition. Biochim. Biophys. Acta http://dx.doi.org/10.1016/j.bbamcr.2012.06.028 (2012)
Berks, B. C., Palmer, T. & Sargent, F. The Tat protein translocation pathway and its role in microbial physiology. Adv. Microb. Physiol. 47, 187–254 (2003)
De Buck, E., Lammertyn, E. & Anne, J. The importance of the twin-arginine translocation pathway for bacterial virulence. Trends Microbiol. 16, 442–453 (2008)
Barkan, A., Miles, D. & Taylor, W. C. Chloroplast gene expression in nuclear, photosynthetic mutants of maize. EMBO J. 5, 1421–1427 (1986)
Settles, A. M. et al. Sec-independent protein translocation by the maize Hcf106 protein. Science 278, 1467–1470 (1997)
Sargent, F. et al. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 17, 3640–3650 (1998)
Bogsch, E. G. et al. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem. 273, 18003–18006 (1998)
Hu, Y., Zhao, E., Li, H., Xia, B. & Jin, C. Solution NMR structure of the TatA component of the twin-arginine protein transport system from gram-positive bacterium Bacillus subtilis. J. Am. Chem. Soc. 132, 15942–15944 (2010)
Berks, B. C. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22, 393–404 (1996)
Chaddock, A. M. et al. A new-type of signal peptide—central role of a twin-arginine motif in transfer signals for the ΔpH-dependent thylakoidal protein translocase. EMBO J. 14, 2715–2722 (1995)
Stanley, N. R., Palmer, T. & Berks, B. C. The twin arginine consensus motif of Tat signal peptides is involved in Sec-independent protein targeting in Escherichia coli. J. Biol. Chem. 275, 11591–11596 (2000)
Alami, M. et al. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol. Cell 12, 937–946 (2003)
Gerard, F. & Cline, K. Efficient twin arginine translocation (Tat) pathway transport of a precursor protein covalently anchored to its initial cpTatC binding site. J. Biol. Chem. 281, 6130–6135 (2006)
Holzapfel, E. et al. The entire N-terminal half of TatC is involved in twin-arginine precursor binding. Biochemistry 46, 2892–2898 (2007)
Kreutzenbeck, P. et al. Escherichia coli twin arginine (Tat) mutant translocases possessing relaxed signal peptide recognition specificities. J. Biol. Chem. 282, 7903–7911 (2007)
Strauch, E. M. & Georgiou, G. Escherichia coli tatC mutations that suppress defective twin-arginine transporter signal peptides. J. Mol. Biol. 374, 283–291 (2007)
Mori, H. & Cline, K. A twin arginine signal peptide and the pH gradient trigger reversible assembly of the thylakoid ΔpH/Tat translocase. J. Cell Biol. 157, 205–210 (2002)
Dabney-Smith, C., Mori, H. & Cline, K. Oligomers of Tha4 organize at the thylakoid Tat translocase during protein transport. J. Biol. Chem. 281, 5476–5483 (2006)
Cline, K. & McCaffery, M. Evidence for a dynamic and transient pathway through the TAT protein transport machinery. EMBO J. 26, 3039–3049 (2007)
Frobel, J., Rose, P. & Muller, M. Early contacts between substrate proteins and TatA translocase component in twin-arginine translocation. J. Biol. Chem. 286, 43679–43689 (2011)
Fritsch, M. J., Krehenbrink, M., Tarry, M. J., Berks, B. C. & Palmer, T. Processing by rhomboid protease is required for Providencia stuartii TatA to interact with TatC and to form functional homo-oligomeric complexes. Mol. Microbiol. 84, 1108–1123 (2012)
Chae, P. S. et al. Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nature Methods 7, 1003–1008 (2010)
Cline, K. & Mori, H. Thylakoid ΔpH-dependent precursor proteins bind to a cpTatC–Hcf106 complex before Tha4-dependent transport. J. Cell Biol. 154, 719–730 (2001)
Punginelli, C. et al. Cysteine scanning mutagenesis and topological mapping of the Escherichia coli twin-arginine translocase TatC Component. J. Bacteriol. 189, 5482–5494 (2007)
Jeong, K. J. et al. A periplasmic fluorescent reporter protein and its application in high-throughput membrane protein topology analysis. J. Mol. Biol. 341, 901–909 (2004)
Behrendt, J., Standar, K., Lindenstrauss, U. & Bruser, T. Topological studies on the twin-arginine translocase component TatC. FEMS Microbiol. Lett. 234, 303–308 (2004)
Drew, D. et al. Rapid topology mapping of Escherichia coli inner-membrane proteins by prediction and PhoA/GFP fusion analysis. Proc. Natl Acad. Sci. USA 99, 2690–2695 (2002)
Gouffi, K., Santini, C. L. & Wu, L. F. Topology determination and functional analysis of the Escherichia coli TatC protein. FEBS Lett. 525, 65–70 (2002)
Kneuper, H. et al. Molecular dissection of TatC defines critical regions essential for protein transport and a TatB-TatC contact site. Mol. Microbiol. 85, 945–961 (2012)
Forrest, L. R., Kramer, R. & Ziegler, C. The structural basis of secondary active transport mechanisms. Biochim. Biophys. Acta 1807, 167–188 (2011)
Buchanan, G. et al. Functional complexity of the twin-arginine translocase TatC component revealed by site-directed mutagenesis. Mol. Microbiol. 43, 1457–1470 (2002)
Mould, R. M. & Robinson, C. A proton gradient is required for the transport of two lumenal oxygen-evolving proteins across the thylakoid membrane. J. Biol. Chem. 266, 12189–12193 (1991)
Zoufaly, S. et al. Mapping precursor-binding site on TatC subunit of twin arginine-specific protein translocase by site-specific photo cross-linking. J. Biol. Chem. 287, 13430–13441 (2012)
Lausberg, F. et al. Genetic evidence for a tight cooperation of TatB and TatC during productive recognition of twin-arginine (Tat) signal peptides in Escherichia coli. PLoS ONE 7, e39867 (2012)
Bolhuis, A., Mathers, J. E., Thomas, J. D., Barrett, C. M. & Robinson, C. TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli. J. Biol. Chem. 276, 20213–20219 (2001)
Tarry, M. J. et al. Structural analysis of substrate binding by the TatBC component of the twin-arginine protein transport system. Proc. Natl Acad. Sci. USA 106, 13284–13289 (2009)
Orriss, G. L. et al. TatBC, TatB, and TatC form structurally autonomous units within the twin arginine protein transport system of Escherichia coli. FEBS Lett. 581, 4091–4097 (2007)
Maldonado, B., Buchanan, G., Muller, M., Berks, B. C. & Palmer, T. Genetic evidence for a TatC dimer at the core of the Escherichia coli twin arginine (Tat) protein translocase. J. Mol. Microbiol. Biotechnol. 20, 168–175 (2011)
Koch, S., Fritsch, M. J., Buchanan, G. & Palmer, T. Escherichia coli TatA and TatB proteins have N-out, C-in topology in intact cells. J. Biol. Chem. 287, 14420–14431 (2012)
Greene, N. P. et al. Cysteine scanning mutagenesis and disulfide mapping studies of the TatA component of the bacterial twin arginine translocase. J. Biol. Chem. 282, 23937–23945 (2007)
Drew, D., Lerch, M., Kunji, E., Slotboom, D. J. & de Gier, J. W. Optimization of membrane protein overexpression and purification using GFP fusions. Nature Methods 3, 303–313 (2006)
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010)
Ashkenazy, H., Erez, E., Martz, E., Pupko, T. & Ben-Tal, N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 (2010)
Tarry, M., Skaar, K., Heijne, G., Draheim, R. R. & Hogbom, M. Production of human tetraspanin proteins in Escherichia coli. Protein Expr. Purif. 82, 373–379 (2012)
Deckert, G. et al. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 392, 353–358 (1998)
Kawate, T. & Gouaux, E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure 14, 673–681 (2006)
Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 (1993)
Winter, G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Cryst. 43, 186–190 (2010)
Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 (2007)
Adams, P. D. et al. The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106 (2011)
Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D 59, 2023–2030 (2003)
Abrahams, J. P. & Leslie, A. G. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D 52, 30–42 (1996)
Bricogne, G. et al. BUSTER 2.11.2 (Global Phasing Ltd., 2011)
Cowtan, K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D 62, 1002–1011 (2006)
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010)
Houtman, J. C. et al. Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: application to adaptor protein complexes in cell signaling. Protein Sci. 16, 30–42 (2007)
Hess, B., Kutzner, C., van der Spoel, D. & Lindahl, E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 (2008)
Monticelli, L., Sorin, E. J., Tieleman, D. P., Pande, V. S. & Colombo, G. Molecular simulation of multistate peptide dynamics: A comparison between microsecond timescale sampling and multiple shorter trajectories. J. Comput. Chem. 29, 1740–1752 (2008)
Scott, K. A. et al. Coarse-grained MD simulations of membrane protein-bilayer self-assembly. Structure 16, 621–630 (2008)
Monticelli, L., Sorin, E. J., Tieleman, D. P., Pande, V. S. & Colombo, G. Molecular simulation of multistate peptide dynamics: a comparison between microsecond timescale sampling and multiple shorter trajectories. J. Comput. Chem. 29, 1740–1752 (2008)
Stansfeld, P. J. & Sansom, M. S. P. From coarse grained to atomistic: a serial multiscale approach to membrane protein simulations. J. Chem. Theory Comput. 7, 1157–1166 (2011)
Acknowledgements
We thank D. Byrne, G. Orriss and R. Owens for their contributions to the early stages of this project; R. Keller for advice; and J. Willem de Gier, D. Daley and R. Huber for providing strains and reagents. This work was supported by the Wellcome Trust (studentships to S.E.R., J.E.G. and M.A.M.; grant 083599, P.R.; grant 092970MA, M.S.P.S.), the Swedish Foundation for Strategic Research (‘Future research leaders 4’ to M.H.), the Swedish Research Council (grant 2010-5061 to M.H.), the E .P. Abrahams Cephalosporin Trust (M.K. and F.R.), the Biotechnology and Biological Sciences Research Council (studentship, M.J.L.; grant BB/E023347/1, S-M.L.; grant BB/1019855/1, P.J.S.), the Medical Research Council (grant G1001640, F.J.; grant G0900888, S.J.), and the European Research Council (Advanced Grant IMPRESS, J.M. and C.V.R.). Work in S.M.L.’s group is funded by the James Martin 21st Century School Vaccine Design Institute.
Author information
Authors and Affiliations
Contributions
The experiments were designed and manuscript written by S.E.R., M.J.T., B.C.B, S.M.L., M.H. and T.P. Experimental work was performed as follows: crystallization trials: S.E.R. and M.J.T.; structure determination: S.E.R., P.R. and S.M.L.; cloning and expression screening: J.E.G., M.J.T., M.J., S-M.L. and M.J.L.; homogeneity screening: M.J.T., M.J., S.E.R., M.A.M. and S.J.; MD simulations: P.J.S. and M.S.P.S; disulphide crosslinking: F.J. and T.P.; signal peptide binding and BN-PAGE: S.E.R., M.K. and F.R.; mass spectrometry: J.M. and C.V.R.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-5, Supplementary References and Supplementary Figures 1-12. (PDF 5169 kb)
Rights and permissions
About this article
Cite this article
Rollauer, S., Tarry, M., Graham, J. et al. Structure of the TatC core of the twin-arginine protein transport system. Nature 492, 210–214 (2012). https://doi.org/10.1038/nature11683
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11683
This article is cited by
-
Strategies of chemolithoautotrophs adapting to high temperature and extremely acidic conditions in a shallow hydrothermal ecosystem
Microbiome (2023)
-
Oligomerization state of the functional bacterial twin-arginine translocation (Tat) receptor complex
Communications Biology (2022)
-
Transport of Folded Proteins by the Tat System
The Protein Journal (2019)
-
Structural Analysis of Tha4, a Twin-arginine Translocase Protein Localized in Plant Thylakoid Membranes
Journal of Plant Biology (2019)
-
Evolution of mitochondrial TAT translocases illustrates the loss of bacterial protein transport machines in mitochondria
BMC Biology (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.