Abstract
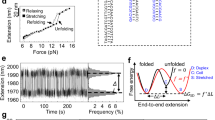
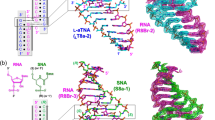
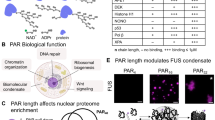
The structure of poly(dA)ṁpoly(dT) currently arouses great interest, mainly because dAnṁdTn stretches are associated with considerable DNA bending1,2. Until recently the heteronomous DNA described by Arnott et al.3, with the poly(dA) and poly(dT) chains in A and B conformations respectively, was the only detailed model of this structure. Following our earlier studies of the interaction of DNA and monovalent ions4–6, we examined the X-ray diffraction of the bivalent Ca2+ salt of poly(dA)-poly(dT) (Ca-poly(d A)ṁpoly(dT)) and found no sign of a heteronomous structure: Ca-poly(dA)ṁpoly(dT) in fibres shows fully equivalent B-type conformations of the opposite sugar–phosphate chains. A revision of the structure of the sodium salt, Na-poly(dA).poly(dT), based on this result, yields only a slightly heteronomous structure with each chain in a B-type conformation, which is in much better agreement with the experimental data underlying the original heteronomous model3. Both structures, Ca- and Na-poly(dA)ṁpoly(dT), have a minor groove narrower than that of the B form: this peculiarity seems to be very important for the interaction of poly(d A)ṁpoly(dT) and biologically significant molecules (including proteins and antibiotics). The specific base-pair positions in poly(dA)ṁpoly(dT) may account for the DNA bending adjacent to dAn-dTn tracts.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Koo, H.-S., Wu, H.-M. & Crothers, D. M. Nature 320, 501–506 (1986).
Hagerman, P. J. Nature 321, 449–450 (1986).
Arnott, S. et al. Nucleic Acids Res. 11, 4141–4145 (1983).
Bartenev, V. N., Golovanov, Eu. I. & Skuratovskii, I. Ya. in Symposium on Biophysics of Nucleic Acids and Nucleoproteins, Tallinn, 179 (Academy of Sciences of the Estonian SSR, Tallinn, 1981).
Bartenev, V. N. et al. J. molec. Biol. 169, 217–234 (1983).
Lipanov, A. A. et al. Biofizika 31, 336–338 (1986).
Langridge, R. et al. J. molec. Biol. 2, 38–64 (1960).
Arnott, S. & Selsing, E. J. molec. Biol. 88, 551–552 (1974).
Smith, P. J. C. & Arnott, S. Acta crystallogr. A34, 3–11 (1978).
Hamilton, W. C. Acta crystallogr. 18, 502–510 (1965).
Fratini, A. V., Kopka, M. L., Drew, H. R. & Dickerson, R. E. J. biol. Chem. 257, 14686–14707 (1982).
Dickerson, R. E., Kopka, M. L. & Pjura, P. in Biological Macromolecules and Assemblies Vol. 2 (eds McPherson, A. & Jurnac, F.) (Wiley, New York, 1985).
Kopka, M. L., Fratini, A. V., Drew, H. R. & Dickerson, R. E. J. molec. Biol. 163, 129–146 (1983).
Alexeev, D. G. et al. in Daresbury Annual Report 1983/84 Appendix, 57 (SERC Daresbury Laboratory, Warrington, 1984).
Skuratovskii, I. Ya., Hasnain, S. S., Alexeev, D. G., Diakun, G. P. & Volkova, L. I. J. inorg. Biochem. (in the press).
Chuprina, V. P. FEBS Lett. 186, 98–102 (1985).
Chuprina, V. P. FEBS Lett. 195, 363–364 (1986).
Chuprina, V. P. Nucleic Acids Res. (in the press).
Ivanov, V. I., Minchenkova, L. E., Minyat, E. E. & Schyolkina, A. K. Cold Spring Harb. Symp. quant. Biol. 47, 243–250 (1983).
Peck, L. J. & Wang, J. C. Nature 292, 375–378 (1981).
Rodes, D. & Klug, A. Nature 292, 378–380 (1981).
Horowitz, D. S. & Wang, J. C. J. molec. Biol. 173, 75–91 (1984).
Wu, H.-M. & Crothers, D. M. Nature 308, 509–513 (1984).
Diekmann, S. & Wang, J. C. J. molec. Biol. 186, 1–11 (1985).
Drew, H. R. & Travers, A. A. Cell 37, 491–502 (1984).
Drew, H. R. & Travers, A. A. J. molec. Biol. 186, 773–790 (1985).
Kopka, M. L., Yoon, C., Goodsell, D., Pjura, P. & Dickerson, R. E. J. molec. Biol. 183, 553–563 (1985).
Dickerson, R. E. & Kopka, M. L. J. biomolec. Struct. Dyn. 3, 423–431 (1985).
Solomon, M. J., Strauss, F. & Varshavsky, A. Proc. natn. Acad. Sci. U.S.A. 83, 1276–1280 (1986).
Bartenev, V. N., Kameneva, N. G. & Lipanov, A. A. Acta crystallogr. (in the press).
Klug, A., Crick, F. H. C. & Wyckoff, H. W. Acta crystallogr. 11, 199–213 (1958).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Alexeev, D., Lipanov, A. & Ya. Skuratovskii, I. Poly(dA)ṁpoly(dT) is a B-type double helix with a distinctively narrow minor groove. Nature 325, 821–823 (1987). https://doi.org/10.1038/325821a0
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/325821a0