Abstract
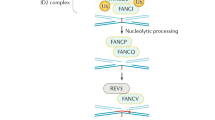
Fanconi anemia (FA) is an autosomal recessive disease with diverse clinical symptoms including developmental anomalies, bone marrow failure and early occurrence of malignancies1. In addition to spontaneous chromosome instability, FA cells exhibit cell cycle disturbances and hypersensitivity to cross-linking agents1. Eight complementation groups (A-H) have been distinguished2, each group possibly representing a distinct FA gene3. The genes mutated in patients of complementation groups A (FANCA; Refs 4,5) and C (FANCC; ref. 6) have been identified, and FANCD has been mapped to chromosome band 3p22-26 (ref. 7). An additional FA gene has recently been mapped to chromosome 9p (ref. 8). Here we report the identification of the gene mutated in group G, FANCG, on the basis of complementation of an FA-G cell line and the presence of pathogenic mutations in four FA-G patients. We identified the gene as human XRCC9, a gene which has been shown to complement the MMC-sensitive Chinese hamster mutant UV40, and is suspected to be involved in DNA post-replication repair or cell cycle checkpoint control9,10. The gene is localized to chromosome band 9p13 (ref. 9), corresponding with a known localization of an FA gene.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Auerbach, A.D., Buchwald, M. & Joenje, H. Fanconi anemia. in The Genetic Basis of Human Cancer (eds Vogelstein, B. & Kinzler, K.W.) 317– 332 (McGraw-Hill, New York, 1998).
Joenje, H. et al. Evidence for at least eight Fanconi anemia genes. Am. J. Hum. Genet. 61, 940–944 (1997).
Buchwald, M. Complementation groups: one or more per gene? Nature Genet. 11, 228–230 (1995).
Lo ten Foe, J.R. et al. Expression cloning of a cDNA for the major Fanconi anemia gene, FAA. Nature Genet. 14, 320– 323 (1996).
The Fanconi anemia/Breast cancer consortium. Positional cloning of Fanconi anemia group A gene. Nature Genet. 14, 324–328 (1996).
Strathdee, C.A., Gavish, H., Shannon, W.R. & Buchwald, M. Cloning of cDNAs for Fanconi's anaemia by functional complementation. Nature 356, 763–767 (1992).
Whitney, M. et al. Microcell mediated chromosome transfer maps the Fanconi anemia group D gene to chromosome 3p. Nature Genet. 11, 341–343 (1995).
Saar, K. et al. Localisation of a Fanconi anaemia gene to chromosome 9p. Eur. J. Hum. Genet. 6, 501–508 (1998).
Liu, N. et al. The human XRCC9 gene corrects chromosomal instability and mutagen sensitivities in CHO UV40 cells. Proc. Natl Acad. Sci. USA 94, 9232–9237 (1997).
Busch, D.B. et al. A CHO mutant, UV40, that is sensitive to diverse mutagens and represents a new complementation group of mitomycin C sensitivity. Mutat. Res. 363, 209–221 (1996).
Ishida, R. & Buchwald, M. Susceptibility of Fanconi's anemia lymphoblasts to DNA cross-linking and alkylating agents. Cancer Res. 42, 4000–4006 (1982).
Schindler, D. & Hoehn, H. Fanconi anemia mutation causes cellular susceptibility to ambient oxygen. Am. J. Hum. Genet. 43, 429–435 (1988).
Kupfer, G.M. et al. The Fanconi anemia proteins, FAA and FAC, interact to form a nuclear complex. Nature Genet. 17, 487–490 (1997).
Hoatlin, M.E. et al. The Fanconi anemia group C gene product is located both in the nucleus and cytoplasm of human cells. Blood 91, 1418–1425 (1998).
Carreau, M. & Buchwald, M. Fanconi's anemia: what have we learned from the genes so far? Mol. Med. Today 4, 201–206 (1998).
Kinzler, K.W. & Vogelstein, B. Gatekeepers and caretakers. Nature 386, 761–763 (1997).
Levran, O. et al. Sequence variation in the Fanconi anemia gene FAA. Proc. Natl Acad. Sci. USA 94, 13051– 13056 (1997).
Auerbach, A.D. & Verlander, P.C. Disorders of DNA replication and repair. Curr. Opin. Pediatr. 9, 600–616 (1997).
Chen, M. et al. Inactivation of Fac in mice produces inducible chromosomal instability and reduced fertility reminiscent of Fanconi anemia. Nature Genet. 12, 448–451 (1996).
Whitney, M.A. et al. Germ cell defects and hematopoietic hypersensitivity to γ interferon in mice with a targeted disruption of the Fanconi anemia C gene. Blood 88, 49–58 (1996).
Strathdee, C.A., Duncan, A.M.V. & Buchwald, M. Evidence for at least four Fanconi anemia genes including FACC on chromosome 9. Nature Genet. 1, 196–198 (1992).
Joenje, H. et al. Classification of Fanconi anemia patients by complementation analysis: evidence for a fifth genetic subtype. Blood 86, 2156–2160 (1995).
Savov, A., Angelicheva, D., Jorfanova, A., Eigel, A. & Kalaydjieva, L. High percentage acrylamide gels improve resolution in SSCP analysis. Nucleic Acids Res. 20, 6741–6742 (1992).
Moshell, A.N., Tarone, R.E., Newfield, S.A., Andrews, A.D. & Robbins, J.H. A simple and rapid method for evaluating the survival of xeroderma pigmentosum lymphoid lines after irradiation with ultraviolet light. In Vitro 17, 299–307 (1981).
Seyschab, H. et al. Comparative evaluation of diepoxybutane sensitivity and cell cycle blockage in the diagnosis of Fanconi anemia. Blood 85, 2233–2237 (1995).
Acknowledgements
We thank R. Dietrich and the FA families who participated in this research project, without whose cooperation this study would have been impossible. We are grateful to B. Schmalenberger, W. Ebell, U. Schulte-Overberg-Schmidt and U. Glöckel for referring patients. We also thank J. Steltenpool and L. van der Weel for technical assistance. This project was supported by the Dutch Cancer Society (project VU-97-1565), the Fanconi Anemia Research Fund Inc., the Commission of the European Union (EUFAR contract PL931562), the National Institutes of Health (HL50131), the Medical Research Council of Canada (MRC) and the Fritz-Thyssen-Stiftung (1997/2061).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
de Winter, J., Waisfisz, Q., Rooimans, M. et al. The Fanconi anaemia group G gene FANCG is identical with XRCC9. Nat Genet 20, 281–283 (1998). https://doi.org/10.1038/3093
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/3093
This article is cited by
-
Fanconi anemia pathway and its relationship with cancer
Genome Instability & Disease (2021)
-
Mechanisms of interstrand DNA crosslink repair and human disorders
Genes and Environment (2016)
-
Update of the human and mouse Fanconi anemia genes
Human Genomics (2015)
-
Coregulation of FANCA and BRCA1 in human cells
SpringerPlus (2014)
-
The structure of the catalytic subunit FANCL of the Fanconi anemia core complex
Nature Structural & Molecular Biology (2010)