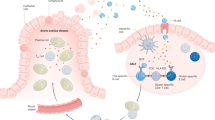
Abstract
Coeliac disease, an inflammatory disease of the small intestine, shares key features with autoimmune disorders, such as susceptibility genes, presence of autoantibodies and T cell-mediated destruction of specific cells. Strikingly, however, continuous exposure to the exogenous dietary antigen gluten and gluten-specific adaptive immunity are required to maintain immunopathology. These observations challenge the notion that autoimmunity requires adaptive immune activation towards self antigens. Using coeliac disease as an example, we propose that other exogenous factors might be identified as drivers of autoimmune processes, in particular when evidence for T cells with specificity for self antigens driving the disease is lacking.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Dicke, W. Coeliac disease. Investigation of the harmful effects of certain types of cereal on patients with coeliac disease. Thesis, Univ. Utrecht (1950).
van Berge-Henegouwen, G. P. & Mulder, C. J. Pioneer in the gluten free diet: Willem-Karel Dicke 1905–1962, over 50 years of gluten free diet. Gut 34, 1473–1475 (1993).
Paulley, J. W. Observation on the aetiology of idiopathic steatorrhoea; jejunal and lymph-node biopsies. Br. Med. J. 2, 1318–1321 (1954).
Green, P. H. & Cellier, C. Celiac disease. N. Engl. J. Med. 357, 1731–1743 (2007).
Dieterich, W. et al. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nature Med. 3, 797–801 (1997).
Sollid, L. M. & Lie, B. A. Celiac disease genetics: current concepts and practical applications. Clin. Gastroenterol. Hepatol. 3, 843–851 (2005).
Trynka, G. et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nature Genet. 43, 1193–1201 (2011).
Smyth, D. J. et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N. Engl. J. Med. 359, 2767–2777 (2008).
Zhernakova, A. et al. Meta-analysis of genome-wide association studies in celiac disease and rheumatoid arthritis identifies fourteen non-HLA shared loci. PLoS Genet. 7, e1002004 (2011).
Trynka, G., Wijmenga, C. & van Heel, D. A. A genetic perspective on coeliac disease. Trends Mol. Med. 16, 537–550 (2010).
Cotsapas, C. et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 7, e1002254 (2011).
Voight, B. F. & Cotsapas, C. Human genetics offers an emerging picture of common pathways and mechanisms in autoimmunity. Curr. Opin. Immunol. 24, 552–557 (2012).
Maurano, M. T. et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 337, 1190–1195 (2012).
Lyons, P. A. et al. Genetically distinct subsets within ANCA-associated vasculitis. N. Engl. J. Med. 367, 214–223 (2012).
van Venrooij, W. J., van Beers, J. J. & Pruijn, G. J. Anti-CCP antibodies: the past, the present and the future. Nature Rev. Rheumatol. 7, 391–398 (2011).
Marzari, R. et al. Molecular dissection of the tissue transglutaminase autoantibody response in celiac disease. J. Immunol. 166, 4170–4176 (2001).
Di Niro, R. et al. High abundance of plasma cells secreting transglutaminase 2-specific IgA autoantibodies with limited somatic hypermutation in celiac disease intestinal lesions. Nature Med. 18, 441–445 (2012).
Bjorck, S., Brundin, C., Lorinc, E., Lynch, K. F. & Agardh, D. Screening detects a high proportion of celiac disease in young HLA-genotyped children. J. Pediatr. Gastroenterol. Nutr. 50, 49–53 (2010).
Klareskog, L., Ronnelid, J., Lundberg, K., Padyukov, L. & Alfredsson, L. Immunity to citrullinated proteins in rheumatoid arthritis. Annu. Rev. Immunol. 26, 651–675 (2008).
Sollid, L. M. Molecular basis of celiac disease. Annu. Rev. Immunol. 18, 53–81 (2000).
Turner, J. R. in Robbins and Cotran Pathologic Basis of Disease (eds Kumar, V., Abbas, A. K., Fausto, N. & Aster, J. C.) 763–831 (Elsevier, 2009).
Hüe, S. et al. A direct role for NKG2D/MICA interaction in villous atrophy during celiac disease. Immunity 21, 367–377 (2004).
Meresse, B. et al. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity 21, 357–366 (2004).
Meresse, B. et al. Reprogramming of CTLs into natural killer-like cells in celiac disease. J. Exp. Med. 203, 1343–1355 (2006).
Jabri, B. & Sollid, L. M. Tissue-mediated control of immunopathology in coeliac disease. Nature Rev. Immunol. 9, 858–870 (2009).
Groh, V., Bruhl, A., El-Gabalawy, H., Nelson, J. L. & Spies, T. Stimulation of T cell autoreactivity by anomalous expression of NKG2D and its MIC ligands in rheumatoid arthritis. Proc. Natl Acad. Sci. USA 100, 9452–9457 (2003).
Ogasawara, K. et al. NKG2D blockade prevents autoimmune diabetes in NOD mice. Immunity 20, 757–767 (2004).
Asquith, D. L. & McInnes, I. B. Emerging cytokine targets in rheumatoid arthritis. Curr. Opin. Rheumatol. 19, 246–251 (2007).
Michalak-Stoma, A. et al. Cytokine network in psoriasis revisited. Eur. Cytokine Netw. 22, 160–168 (2011).
Rentzos, M. & Rombos, A. The role of IL-15 in central nervous system disorders. Acta Neurol. Scand. 125, 77–82 (2012).
Mention, J. J. et al. Interleukin 15: a key to disrupted intraepithelial lymphocyte homeostasis and lymphomagenesis in celiac disease. Gastroenterology 125, 730–745 (2003).
Tang, F. et al. Cytosolic PLA2 is required for CTL-mediated immunopathology of celiac disease via NKG2D and IL-15. J. Exp. Med. 206, 707–719 (2009).
Groh, V. et al. Costimulation of CD8αβ T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nature Immunol. 2, 255–260 (2001).
Roberts, A. I. et al. NKG2D receptors induced by IL-15 costimulate CD28-negative effector CTL in the tissue microenvironment. J. Immunol. 167, 5527–5530 (2001).
Sulkanen, S. et al. Tissue transglutaminase autoantibody enzyme-linked immunosorbent assay in detecting celiac disease. Gastroenterology 115, 1322–1328 (1998).
Bardella, M. T. et al. Coeliac disease: a histological follow-up study. Histopathology 50, 465–471 (2007).
Molberg, O. et al. Gliadin specific, HLA DQ2-restricted T cells are commonly found in small intestinal biopsies from coeliac disease patients, but not from controls. Scand. J. Immunol. 46, 103–108 (1997).
Sollid, L. M., Qiao, S. W., Anderson, R. P., Gianfrani, C. & Koning, F. Nomenclature and listing of celiac disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules. Immunogenetics 64, 455–460 (2012).
Lundin, K. E. et al. Gliadin-specific, HLA-DQ(α1*0501, β1*0201) restricted T cells isolated from the small intestinal mucosa of celiac disease patients. J. Exp. Med. 178, 187–196 (1993).
Lundin, K. E., Scott, H., Fausa, O., Thorsby, E. & Sollid, L. M. T cells from the small intestinal mucosa of a DR4, DQ7/DR4, DQ8 celiac disease patient preferentially recognize gliadin when presented by DQ8. Hum. Immunol. 41, 285–291 (1994).
Molberg, O. et al. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nature Med. 4, 713–717 (1998).
van de Wal, Y. et al. Selective deamidation by tissue transglutaminase strongly enhances gliadin-specific T cell reactivity. J. Immunol. 161, 1585–1588 (1998).
Tollefsen, S. et al. HLA-DQ2 and -DQ8 signatures of gluten T cell epitopes in celiac disease. J. Clin. Invest. 116, 2226–2236 (2006).
Osman, A. A. et al. B cell epitopes of gliadin. Clin. Exp. Immunol. 121, 248–254 (2000).
Shan, L. et al. Structural basis for gluten intolerance in celiac sprue. Science 297, 2275–2279 (2002).
Bodd, M. et al. HLA-DQ2-restricted gluten-reactive T cells produce IL-21 but not IL-17 or IL-22. Mucosal Immunol. 3, 594–601 (2010).
Abadie, V., Sollid, L. M., Barreiro, L. B. & Jabri, B. Integration of genetic and immunological insights into a model of celiac disease pathogenesis. Annu. Rev. Immunol. 29, 493–525 (2011).
Sollid, L. M. Coeliac disease: dissecting a complex inflammatory disorder. Nature Rev. Immunol. 2, 647–655 (2002).
Mäki, M. in Seventh International Symposium on Coeliac Disease (eds Feighery, C. & O'Farelly, C.) 246–252 (Oak Tree Press, 1992).
Sollid, L. M., Molberg, O., McAdam, S. & Lundin, K. E. Autoantibodies in coeliac disease: tissue transglutaminase—guilt by association? Gut 41, 851–852 (1997).
Fleckenstein, B. et al. Molecular characterization of covalent complexes between tissue transglutaminase and gliadin peptides. J. Biol. Chem. 279, 17607–17616 (2004).
Prause, C. et al. Antibodies against deamidated gliadin as new and accurate biomarkers of childhood coeliac disease. J. Pediatr. Gastroenterol. Nutr. 49, 52–58 (2009).
Devendra, D. & Eisenbarth, G. S. Interferon alpha—a potential link in the pathogenesis of viral-induced type 1 diabetes and autoimmunity. Clin. Immunol. 111, 225–233 (2004).
Fujinami, R. S. & Oldstone, M. B. Amino acid homology between the encephalitogenic site of myelin basic protein and virus: mechanism for autoimmunity. Science 230, 1043–1045 (1985).
Wucherpfennig, K. W. & Strominger, J. L. Molecular mimicry in T cell-mediated autoimmunity: viral peptides activate human T cell clones specific for myelin basic protein. Cell 80, 695–705 (1995).
Lawson, C. M. Evidence for mimicry by viral antigens in animal models of autoimmune disease including myocarditis. Cell. Mol. Life Sci. 57, 552–560 (2000).
Pabst, O. & Mowat, A. M. Oral tolerance to food protein. Mucosal Immunol. 5, 232–239 (2012).
DePaolo, R. W. et al. Co-adjuvant effects of retinoic acid and IL-15 induce inflammatory immunity to dietary antigens. Nature 471, 220–224 (2011).
Monteleone, G. et al. Role of interferon α in promoting T helper cell type 1 responses in the small intestine in coeliac disease. Gut 48, 425–429 (2001).
Raki, M. et al. Plasmacytoid dendritic cells are scarcely represented in the human gut mucosa and are not recruited to the celiac lesion. Mucosal Immunol. 23 Jan 2013 (doi:10.1038/mi.2012.136)
Medzhitov, R. & Janeway, C. A. Jr. Innate immune recognition and control of adaptive immune responses. Semin. Immunol. 10, 351–353 (1998).
Di Sabatino, A. et al. Evidence for the role of interferon-alfa production by dendritic cells in the Th1 response in celiac disease. Gastroenterology 133, 1175–1187 (2007).
Maiuri, L. et al. Association between innate response to gliadin and activation of pathogenic T cells in coeliac disease. Lancet 362, 30–37 (2003).
Barone, M. V. et al. Growth factor-like activity of gliadin, an alimentary protein: implications for coeliac disease. Gut 56, 480–488 (2007).
Terrazzano, G. et al. Gliadin regulates the NK–dendritic cell cross-talk by HLA-E surface stabilization. J. Immunol. 179, 372–381 (2007).
Junker, Y. et al. Wheat amylase trypsin inhibitors drive intestinal inflammation via activation of toll-like receptor 4. J. Exp. Med. 209, 2395–2408 (2012).
Troncone, R. & Auricchio, S. Rotavirus and celiac disease: clues to the pathogenesis and perspectives on prevention. J. Pediatr. Gastroenterol. Nutr. 44, 527–528 (2007).
Stene, L. C. et al. Rotavirus infection frequency and risk of celiac disease autoimmunity in early childhood: a longitudinal study. Am. J. Gastroenterol. 101, 2333–2340 (2006).
Hober, D. & Sauter, P. Pathogenesis of type 1 diabetes mellitus: interplay between enterovirus and host. Nature Rev. Endocrinol. 6, 279–289 (2010).
Siegel, M. et al. Extracellular transglutaminase 2 is catalytically inactive, but is transiently activated upon tissue injury. PLoS ONE 3, e1861 (2008).
Jun, H. S. & Yoon, J. W. A new look at viruses in type 1 diabetes. Diabetes Metab. Res. Rev. 19, 8–31 (2003).
Rizza, P., Moretti, F. & Belardelli, F. Recent advances on the immunomodulatory effects of IFN-α: implications for cancer immunotherapy and autoimmunity. Autoimmunity 43, 204–209 (2010).
Carrero, J. A., Calderon, B. & Unanue, E. R. Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. J. Exp. Med. 200, 535–540 (2004).
Woodward, J. J., Iavarone, A. T. & Portnoy, D. A. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328, 1703–1705 (2010).
Manca, C. et al. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-α/β. Proc. Natl Acad. Sci. USA 98, 5752–5757 (2001).
Jones, J. W. et al. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl Acad. Sci. USA 107, 9771–9776 (2010).
Round, J. L. & Mazmanian, S. K. The gut microbiota shapes intestinal immune responses during health and disease. Nature Rev. Immunol. 9, 313–323 (2009).
Maynard, C. L., Elson, C. O., Hatton, R. D. & Weaver, C. T. Reciprocal interactions of the intestinal microbiota and immune system. Nature 489, 231–241 (2012).
Hooper, L. V., Littman, D. R. & Macpherson, A. J. Interactions between the microbiota and the immune system. Science 336, 1268–1273 (2012).
Atkinson, M. A. & Chervonsky, A. Does the gut microbiota have a role in type 1 diabetes? Early evidence from humans and animal models of the disease. Diabetologia 55, 2868–2877 (2012).
Mathis, D. & Benoist, C. Microbiota and autoimmune disease: the hosted self. Cell Host Microbe 10, 297–301 (2011).
Forsberg, G. et al. Presence of bacteria and innate immunity of intestinal epithelium in childhood celiac disease. Am. J. Gastroenterol. 99, 894–904 (2004).
Tjellstrom, B. et al. Gut microflora associated characteristics in children with celiac disease. Am. J. Gastroenterol. 100, 2784–2788 (2005).
Sanz, Y. et al. Differences in faecal bacterial communities in coeliac and healthy children as detected by PCR and denaturing gradient gel electrophoresis. FEMS Immunol. Med. Microbiol. 51, 562–568 (2007).
Di Cagno, R. et al. Duodenal and faecal microbiota of celiac children: molecular, phenotype and metabolome characterization. BMC Microbiol. 11, 219 (2011).
Nistal, E. et al. Differences of small intestinal bacteria populations in adults and children with/without celiac disease: effect of age, gluten diet, and disease. Inflamm. Bowel Dis. 18, 649–656 (2012).
De Palma, G. et al. Intestinal dysbiosis and reduced immunoglobulin-coated bacteria associated with coeliac disease in children. BMC Microbiol. 10, 63 (2010).
Collado, M. C., Donat, E., Ribes-Koninckx, C., Calabuig, M. & Sanz, Y. Specific duodenal and faecal bacterial groups associated with paediatric coeliac disease. J. Clin. Pathol. 62, 264–269 (2009).
Nadal, I., Donat, E., Ribes-Koninckx, C., Calabuig, M. & Sanz, Y. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J. Med. Microbiol. 56, 1669–1674 (2007).
Sanchez, E. et al. Reduced diversity and increased virulence-gene carriage in intestinal enterobacteria of coeliac children. BMC Gastroenterol. 8, 50 (2008).
Sanchez, E., Ribes-Koninckx, C., Calabuig, M. & Sanz, Y. Intestinal Staphylococcus spp. and virulent features associated with coeliac disease. J. Clin. Pathol. 65, 830–834 (2012).
Kagnoff, M. F. et al. Evidence for the role of a human intestinal adenovirus in the pathogenesis of coeliac disease. Gut 28, 995–1001 (1987).
Benoist, C. & Mathis, D. Autoimmunity provoked by infection: how good is the case for T cell epitope mimicry? Nature Immunol. 2, 797–801 (2001).
Wegner, N. et al. Peptidylarginine deiminase from Porphyromonas gingivalis citrullinates human fibrinogen and α-enolase: implications for autoimmunity in rheumatoid arthritis. Arthritis Rheum. 62, 2662–2672 (2010).
Wu, H. J. et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827 (2010).
Saleh, M. & Elson, C. O. Experimental inflammatory bowel disease: insights into the host-microbiota dialog. Immunity 34, 293–302 (2011).
Khan, K. J. et al. Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am. J. Gastroenterol. 106, 661–673 (2011).
Janowitz, H. D., Croen, E. C. & Sachar, D. B. The role of the fecal stream in Crohn's disease: an historical and analytic review. Inflamm. Bowel Dis. 4, 29–39 (1998).
Mattner, J. et al. Liver autoimmunity triggered by microbial activation of natural killer T cells. Cell Host Microbe 3, 304–315 (2008).
Shewry, R. P., Tatham, A. & Kasarda, D. D. in Coeliac disease (ed. Marsh, M. N.) (Blackwell Scientific Publications, 1992).
Alp, M. H. & Wright, R. Autoantibodies to reticulin in patients with idiopathic steatorrhoea, coeliac disease, and Crohn's disease, and their relation to immunoglobulins and dietary antibodies. Lancet 2, 682–685 (1971).
Seah, P. P., Fry, L., Rossiter, M. A., Hoffbrand, A. V. & Holborow, E. J. Anti-reticulin antibodies in childhood coeliac disease. Lancet 2, 681–682 (1971).
Chorzelski, T. P. et al. IgA anti-endomysium antibody. A new immunological marker of dermatitis herpetiformis and coeliac disease. Br. J. Dermatol. 111, 395–402 (1984).
Ladinser, B., Rossipal, E. & Pittschieler, K. Endomysium antibodies in coeliac disease: an improved method. Gut 35, 776–778 (1994).
Sanchez, D. et al. Occurrence of IgA and IgG autoantibodies to calreticulin in coeliac disease and various autoimmune diseases. J. Autoimmun. 15, 441–449 (2000).
Clemente, M. G. et al. Enterocyte actin autoantibody detection: a new diagnostic tool in celiac disease diagnosis: results of a multicenter study. Am. J. Gastroenterol. 99, 1551–1556 (2004).
Rostom, A. et al. The diagnostic accuracy of serologic tests for celiac disease: a systematic review. Gastroenterology 128, S38–S46 (2005).
Husby, S. et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J. Pediatr. Gastroenterol. Nutr. 54, 136–160 (2012).
Lorand, L. & Graham, R. M. Transglutaminases: crosslinking enzymes with pleiotropic functions. Nature Rev. Mol. Cell. Biol. 4, 140–156 (2003).
Piper, J. L., Gray, G. M. & Khosla, C. High selectivity of human tissue transglutaminase for immunoactive gliadin peptides: implications for celiac sprue. Biochemistry 41, 386–393 (2002).
Stamnaes, J., Pinkas, D. M., Fleckenstein, B., Khosla, C. & Sollid, L. M. Redox regulation of transglutaminase 2 activity. J. Biol. Chem. 285, 25402–25409 (2010).
Dorum, S. et al. The preferred substrates for transglutaminase 2 in a complex wheat gluten digest are peptide fragments harboring celiac disease T-cell epitopes. PLoS ONE 5, e14056 (2010).
Maslowski, K. M. & Mackay, C. R. Diet, gut microbiota and immune responses. Nature Immunol. 12, 5–9 (2011).
Acknowledgements
The work was supported by grants from the Research Council of Norway, the European Research Council and the South-East Norway Regional Health Authority to L.M.S. and by grants from the US National Institutes of Health (grants RO1DK063158, RO1DK58727and P30DK42086) to B.J.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Related links
Rights and permissions
About this article
Cite this article
Sollid, L., Jabri, B. Triggers and drivers of autoimmunity: lessons from coeliac disease. Nat Rev Immunol 13, 294–302 (2013). https://doi.org/10.1038/nri3407
Published:
Issue Date:
DOI: https://doi.org/10.1038/nri3407
This article is cited by
-
Primary sclerosing cholangitis (PSC) and inflammatory bowel disease (IBD): a condition exemplifying the crosstalk of the gut–liver axis
Experimental & Molecular Medicine (2023)
-
Characterizations of a neutralizing antibody broadly reactive to multiple gluten peptide:HLA-DQ2.5 complexes in the context of celiac disease
Nature Communications (2023)
-
In a large Juvenile Idiopathic Arthritis (JIA) cohort, concomitant celiac disease is associated with family history of autoimmunity and a more severe JIA course: a retrospective study
Pediatric Rheumatology (2022)
-
Single cell transcriptomic analysis of the immune cell compartment in the human small intestine and in Celiac disease
Nature Communications (2022)
-
Review on pediatric coeliac disease from a clinical perspective
European Journal of Pediatrics (2022)