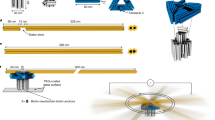
Abstract
The biophysical and biochemical properties of motor proteins have been well-studied, but these motors also show promise as mechanical components in hybrid nano-engineered systems1,2,3,4. The cytoplasmic F1 fragment of the adenosine triphosphate synthase (F1-ATPase) can function as an ATP-fuelled rotary motor4,5,6,7 and has been integrated into self-assembled nanomechanical systems as a mechanical actuator4,8. Here we present the rational design, construction and analysis of a mutant F1-ATPase motor containing a metal-binding site that functions as a zinc-dependent, reversible on/off switch. Repeated cycles of zinc addition and removal by chelation result in inhibition and restoration, respectively, of both ATP hydrolysis and motor rotation of the mutant, but not of the wild-type F1 fragment. These results demonstrate the ability to engineer chemical regulation into a biomolecular motor and represent a critical step towards controlling integrated nanomechanical devices at the single-molecule level.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hess, H., Clemmens, J., Qin, D., Howard, J. & Vogel, V. Light-controlled molecular shuttles made from motor proteins carrying cargo on engineered surfaces. Nanoletters 1, 235–239 (2001).
Bohm, K.J., Stracke, R., Muhlig, P. & Unger, E. Motor protein-driven unidirectional transport of micrometer-sized cargoes across isopolar microtubule arrays. Nanotechnology 12, 238–244 (2001).
Hiratsuka, Y., Tada, T., Oiwa, K., Kanayama, T. & Uyeda, T. Controlling the direction of kinesin-driven microtubule movements along microlithographic tracks. Biophys. J. 81, 1555–1561 (2001).
Soong, R.K. et al. Powering an inorganic nanodevice with a biomolecular motor. Science 290, 1555–1558 (2000).
Noji, H., Yasuda, R., Yoshida, M. & Kinosita, K. Direct observation of the rotation of F1-ATPase. Nature 386, 299–302 (1997).
Yasuda, R., Noji, H., Kinosita, K. & Yoshida, M. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120 degrees steps. Cell 93, 1117–1124 (1998).
Yasuda, R., Noji, H., Yoshida, M., Kinosita, K. & Itoh, H. Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410, 898–904 (2001).
Soong, R.K., Neves, H.P., Bachand, G.D., Schmidt, J. & Montemagno, C.D. Engineering hybrid nanoscale devices powered by biomolecular motors. Biomed. Microdevices 3, 71–73 (2001).
Abrahams, P., Leslie, A.G.W., Lutter, R. & Walker, J.E. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 (1994).
Perutz, M.F. Mechanism of cooperativity and allosteric regulation in proteins. Q. Rev. Biophys. 22, 139–237 (1989).
Glusker, J.P. Structural aspects of metal liganding to functional groups in proteins. Adv. Prot. Chem. 42, 1–76 (1991).
Hellinga, H.W. & Richards, F.M. Construction of new ligand-binding sites in proteins of known structure. 1. Computer-aided modeling of sites with predefined geometry. J. Mol. Biol. 222, 763–785 (1991).
Marvin, J.S. & Hellinga, H.W. Conversion of a maltose receptor into a zinc biosensor by computational design. Proc. Natl Acad. Sci. USA 98, 4955–4960 (2001).
Benson, D., Haddy, A.E. & Hellinga, H.W. Converting a maltose receptor into a nascent binuclear copper oxygenase by computational design. Biochemistry 41, 3262–3269 (2002).
Marvin, J. et al. The rational design of allosteric interactions in a monomeric protein and its applications to the construction of biosensors. Proc. Natl Acad. Sci. USA 98, 4366–4371 (1997).
Shirakihara, Y. et al. The crystal structure of the nucleotide-free subcomplex of F1-ATPase from the thermophilic Bacillus PS3 is a symmetric trimer. Structure 5, 825–836 (1997).
Bachand, G.D. & Montemagno, C.D. Constructing organic/inorganic NEMS devices powered by biomolecular motors. Biomed. Microdevices 2, 179–184 (2000).
Pullman, M.E., Penefsky, H.S., Datta, A. & Racker, E. Partial resolution of the enzyme catalyzing oxidative phosphorylation. I. Purification and properties of soluble, dinitrophenol-stimulated adenosine triphosphatase. J. Biol. Chem. 235, 3322–3329 (1960).
Segel, I.H. Enzyme Kinetics (Wiley, New York, 1975).
Hunt, A.J., Gittes, F. & Howard, J. The force exerted by a single kinesin molecule against a viscous load. Biophys. J. 67, 766–781 (1994).
Acknowledgements
We thank C. Xu for help in the protein modelling of the TF1 structure. This research was supported in part by grants from NASA (NAG5-8775), DARPA (N00014-99-1-0436) and NSF (ECS-0084732). Sandia is a multiprogram laboratory operated by Sandia Corporation, a Lockheed Martin Company, for the US Department of Energy under contract DE-AC04-94AL85000.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Liu, H., Schmidt, J., Bachand, G. et al. Control of a biomolecular motor-powered nanodevice with an engineered chemical switch. Nature Mater 1, 173–177 (2002). https://doi.org/10.1038/nmat761
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmat761
This article is cited by
-
Design of allosteric sites into rotary motor V1-ATPase by restoring lost function of pseudo-active sites
Nature Chemistry (2023)
-
Reconstitution of FoF1-ATPase-based biomimetic systems
Nature Reviews Chemistry (2019)
-
Creating biomolecular motors based on dynein and actin-binding proteins
Nature Nanotechnology (2017)
-
Engineering controllable bidirectional molecular motors based on myosin
Nature Nanotechnology (2012)
-
Two-stimuli manipulation of a biological motor
Journal of Nanobiotechnology (2009)