Abstract
Omi, also known as high temperature requirement factor A2 (HtrA2), is a serine protease that was originally identified as a proapoptotic protein. Like Smac/Diablo, it antagonizes inhibitor of apoptosis proteins when released into the cytosol on apoptotic stimulation. Loss of its protease activity in mnd2 (motor neuron degeneration 2) mice is associated with neurodegeneration. However, the detailed mechanisms by which Omi regulates the pathogenesis of neurodegenerative disease remain largely unknown. We report here that Omi participates in the pivotal cellular degradation process known as autophagy. It activates autophagy through digestion of Hax-1, a Bcl-2 family-related protein that represses autophagy in a Beclin-1 (mammalian homologue of yeast ATG6)-dependent pathway. Moreover, Omi-induced autophagy facilitates the degradation of neurodegenerative proteins such as pathogenic A53T α-synuclein and truncated polyglutamine-expanded huntingtin, as well as the endogenous autophagy substrate p62. Knockdown of Omi decreases the basal level of autophagy and increases the level of the above target proteins. Furthermore, S276C Omi, the protease-defective mutant found in mnd2 mice, fails to regulate autophagy. Increased autophagy substrates and the formation of aggregate structures are observed in the brains of mnd2 mice. These results identify Omi as a novel regulator of autophagy and suggest that Omi might be important in the cellular quality control of proteins involved in neurodegenerative diseases.
Similar content being viewed by others
Main
Omi/HtrA2 belongs to the high-temperature requirement factor A (HtrA) family. HtrA was originally identified as a heat-shock-induced serine protease in Escherichia coli that degrades misfolded proteins.1 HtrA proteases contain one or two C-terminal PDZ domains that activate the trypsin-like protease domain once they recognize misfolded or damaged proteins.2 The mammalian HtrA2 protease, Omi, is much better known than other HtrA family members for its proapoptotic activity.3 Most Omi resides in the intermembrane space of the mitochondria; however, endoplasmic reticulum (ER) and Golgi localization have also been reported.4 Under apoptotic stimulation, a mature processed form of Omi is released into the cytosol where it binds to the cytosolic inhibitor of apoptosis proteins (IAPs) and inactivates their caspase-inhibitory activity5 by direct cleavage.3 Mature Omi that lacks the mitochondrial targeting sequence induces dramatic apoptosis-like morphological changes and cell death that is not blocked by caspase inhibitors.6
Recently, increasing evidence has linked Omi to neurodegeneration and the cellular protein quality control system. Motor neuron degeneration 2 (mnd2) homozygous mice, in which the missense mutation S276C in Omi leads to a remarkable loss of Omi protease activity, showed severe neurodegeneration in the striatum and less severe neurodegeneration in the brain stem and spinal cord.7 Missense mutations in the gene coding for Omi were reported to be associated with Parkinson's disease (PD).8 In agreement with these findings, accumulation of Omi was found in neuronal and glial inclusions in brains with α-synucleinopathies,9 as well as in Lewy bodies.8 Omi knockout (KO) mice suffered the loss of a population of neurons in the striatum, which resulted in a neurodegenerative disorder with a PD phenotype. These mice died within 30 days after birth, similarly to mnd2 mice.10 Furthermore, many other neurodegenerative disease proteins, such as presenilin-14, 11 or amyloid precursor protein,12 were reported to be associated with Omi.
Autophagy (from the Greek ‘auto’ for ‘self’ and ‘phagy’ for ‘eat’) is a protein degradation process that digests intracellular organelles and proteins in response to nutritional deprivation or other stimuli such as ER stress. Under certain conditions, autophagy is activated for avoidance of cell death or suppression of apoptosis, whereas in other circumstances, it constitutes an alternative cell death pathway called autophagic cell death or type II cell death. The dead cells appear to be round and detached, and they have a convoluted plasma membrane permeable to vital dyes.13 Notably, these features share similarities with mature Omi-induced cell death.6 Moreover, defective autophagy has been frequently implicated in neurodegenerative disorders because of the accumulation of aberrant structures and aggregated proteins in neurons. For instance, conditional KO experiments revealed that animals lacking ATG5 or ATG7 (two essential autophagy genes) suffered from neurodegeneration in the central nervous system, indicating that autophagy is indispensable for neuronal cells.14, 15
Here, we investigated the role Omi-mediated regulation of autophagic activity in the degradation of proteins involved in neurodegenerative diseases.
Results
Omi activates autophagy in a dose-dependent manner
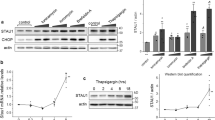
Because extramitochondrially expressed mature Omi promotes both caspase-dependent and -independent cell death,6 we investigated whether autophagic cell death, so-called type II cell death, is involved. HEK293 cells were transfected with mature Omi containing C-terminal MYC or HA tags. Overexpression of mature Omi induced significant cell death, as evidenced by condensed nuclei in HEK293 cells (Supplementary Figure S1a, upper panel). Meanwhile, mature Omi increased the turnover of LC3-I to LC3-II, a PE-conjugated form of microtubule-associated protein 1 light chain 3 (LC3) that is widely used as an autophagy marker (Figure 1a). Next, we examined whether non-proapoptotic full-length Omi (referred to hereafter as Omi) enhances autophagy. In HEK293 cells, Omi increased LC3-II levels, although no apoptosis was induced (Figure 1b, Supplementary Figure S1a and b). ATG5 is well known as an autophagy essential gene.16 We examined the effects of Omi on autophagy activation in ATG5 wild-type (WT) and KO mouse embryonic fibroblast (MEF) cells. Omi increased LC3-II levels in ATG5 WT cells, but not in autophagy-deficient ATG5 KO cells (Figure 1c).
Induction of autophagy by Omi. (a) Mature Omi (M-Omi) induces autophagy. HEK293 cells transfected with M-Omi-MYC were collected for immunoblot analysis using anti-Omi or LC3 antibodies. Endo Omi, endogenous Omi. (b) Full-length Omi triggers autophagy. A similar experiment as in (a) was carried out using Omi-MYC. (c) Omi fails to trigger autophagy in ATG5 KO MEF cells. (d) For TEM analysis, HeLa cells transfected with mock HA empty vector (upper panel, left) or Omi-HA (two panels on the right) were processed for TEM analysis. Rapamycin-treated cells were used as a positive control (lower panel, left). Double-membrane autophagic vesicles are indicated with black arrows, and autolysosome is indicated with white arrow. (e) Representative micrograph showing redistribution of EGFP-LC3 into puncta by Omi-HA. HeLa cells were transfected with EGFP-LC3 and mock vector or Omi-HA plasmid and subjected to immunofluorescence analysis 48 h after transfection. (f) The percentage of cells showing >5 intense LC3 puncta in (e) was quantified (mean±S.E.M., n=3; *P<0.05 one-way ANOVA). (g) Omi induces autophagy in a concentration-dependent manner. Increasing amounts of Omi-MYC were transfected into a HeLa cell line stably expressing EGFP-LC3, and the cell lysates were subjected to immunoblotting 48 h after transfection. The bands of free EGFP were arrowed. (h) Wortmannin inhibits the autophagy triggered by Omi in HeLa cells
The presence of double-membrane vesicle structures, that is, autophagosomes and autolysosomes, is a hallmark of autophagy. We evaluated the formation of these structures in Omi-transfected cells using transmission electron microscopy (TEM). Omi and rapamycin, an inducer of autophagy through the inactivation of mTOR, increased the formation of typical double-membrane autophagosomes (Figure 1d). There were few autophagosomes in control cells transfected with mock vector alone (Figure 1d, upper panel). However, in the rapamycin-treated cells (Figure 1d, middle panel) or Omi-overexpressing cells (Figure 1d, lower panel), numerous typical autophagosomes and autolysosomes were observed.
EGFP-LC3 has been widely used to monitor autophagy, as measured by an increase in punctate LC3 signal, because it is incorporated into autophagosomes during autophagy.17 Redistribution of EGFP-LC3 into puncta was observed in Omi-transfected cells, but not in Omi nontransfected cells (Figure 1e and f). When autophagy is activated, the fusion protein EGFP-LC3 is delivered to the lysosome; the LC3 portion of the chimera is sensitive to degradation, whereas the EGFP protein is relatively resistant to hydrolysis. Therefore, the appearance and the amount of free EGFP observed on an immunoblot can be used to monitor the lysis of the inner autophagosomal membrane and breakdown of the cargo, thereby permitting evaluation of autophagy flux.17 In a HeLa cell line stably expressing EGFP-LC3, transfection with increased amounts of Omi resulted in an increase of free EGFP release (Figure 1g).
To investigate whether Omi-induced autophagy could be inhibited by autophagy inhibitors, we treated Omi-transfected cells with wortmannin (WM).17 WM significantly decreased the level of LC3-II in Omi-transfected cells (Figure 1h).
Regulation of basal level and stress-induced autophagy by endogenous Omi
Constitutive basal autophagy is essential for systematic quality control and cellular homeostasis. Maintenance of autophagy at a basal level in most cells contributes to the routine turnover of cytoplasmic components, and autophagy is rapidly upregulated when cells need to generate intracellular nutrients and energy. As previously reported, ER stress can activate autophagy18 and increase Omi expression4 (confirmed in Supplementary Figure S2a), indicating that endogenous Omi might be involved in regulating constitutive basal autophagy. We therefore examined whether endogenous Omi levels were related to stress-induced autophagy activation. Synchronous increases in autophagy and Omi levels were observed in WT MEF cells treated with tunicamycin, an ER stress drug (Figure 2a). However, in ATG5 KO MEF cells, no LC3-II band was detected, although Omi was upregulated (Figure 2a, right panel).
Regulation of basal level and stress-induced autophagy by endogenous Omi. (a) Synchronous upregulation of Omi and LC3-II level on ER stress in ATG5 WT, but not in ATG5 KO MEF cells. WT MEF or ATG5 KO MEF cells were treated with DMSO or increasing levels of tunicamycin (0.5–1 μg/ml) for 8 h at 37°C and subjected to immunoblot analysis. (b) Knockdown of Omi decreases basal autophagy, which is restored by the same sense mutants of WT Omi-HA (designed to be resistant to small-interfering RNAs against Omi), but not the same sense mutants of S276C Omi-HA in HeLa cells. (c, d) Decreased autophagic activation in response to starvation (HBSS, 2 h) and tunicamycin (0.5 μg/ml, 6 h) by knockdown of Omi in a HeLa EGFP-LC3 stable cell line. In (c), representative microphotographs are shown. The percentage of cells with EGFP-LC3 puncta is quantitated in (d) (mean±S.E.M., n=3, *P<0.05 one-way ANOVA). (e) Formation of abnormal autophagic vesicles in Omi knockdown HeLa cells in response to ER stress
We further assessed the effects of endogenous Omi on autophagy using small-interfering RNA (siRNA)-mediated silencing to knock down Omi expression. The knockdown efficiencies of two distinct siRNAs against Omi are shown in Supplementary Figure S2b. The LC3-II level was decreased in Omi knockdown cells compared with negative control cells (Figure 2b). We next generated Omi siRNA-resistant same sense mutants (SSmutant) of WT Omi and S276C Omi. The loss of protease function in the S276C Omi was confirmed using its known substrate β-casein (Supplementary Figure S2c). Retransfection of a noninterferable cDNA coding for WT Omi, but not its protease-inactive mutant (S276C), recovered LC3-II level in Omi knockdown cells (Figure 2b).
We next examined the formation of punctate LC3 in Omi-silenced HeLa cells stably expressing EGFP-LC3 under nutrient deprivation or ER stress. Knockdown of Omi decreased the formation of punctate EGFP-LC3 induced by starvation and ER stress (Figure 2c and d). In addition, abnormality of the autophagic vesicles (destructive or unclosed membrane) was observed in Omi knockdown cells after tunicamycin treatment (Figure 2e, lower panel). These data collectively support the notion that endogenous Omi is involved in the regulation of autophagy.
Enhanced clearance of neurodegenerative disease proteins by Omi-induced autophagy
The clearance of aggregation-prone proteins like mutant huntingtin (htt) fragments, other polyglutamine (polyQ) proteins, mutant α-synuclein and tau is strongly dependent on autophagy.19, 20 We examined the ability of Omi-induced autophagy to clear such neurodegenerative disease proteins. Using autophagy-inducing or -inhibiting drugs, we confirmed that A53T α-synuclein is an autophagy substrate (Supplementary Figure S3a). We next transfected HEK293 cells with MYC-tagged Omi along with A53T α-synuclein-EGFP or tHtt-Q60-EGFP (a truncated form of htt with 60 polyQs).21 The amounts of A53T α-synuclein-EGFP and tHtt-Q60-EGFP were decreased in cells co-transfected with Omi, whereas in cells co-transfected with Omi and EGFP, the amount of EGFP was not changed (Figure 3a and b).
Facilitation of the clearance of autophagy substrates by Omi. (a) Accelerated clearance of autophagy substrates by Omi. EGFP, tHtt-Q60-EGFP or A53T α-synuclein-EGFP levels in HEK293 cells transfected with or without Omi were detected using an anti-GFP antibody. (b) Quantitative analysis of the relative density compared to loading controls (endogenous Omi) for protein levels from (a). Mean±S.E.M., n=3, *P<0.05, one-way ANOVA. (c) Omi facilitates the clearance of A53T α-synuclein-EGFP in an N2a A53T α-synuclein-EGFP stable cell line. (d) Quantitative analysis of the relative density compared with loading controls (endogenous Omi) from (c). The data represent mean±S.E.M., n=3, *P<0.05, one-way ANOVA. (e) Omi increases degradation of A53T α-synuclein in ATG5 WT but not in ATG5 KO MEF cells. (f) No influence of the proteasome inhibitor MG132 (5 μM 18 h) on Omi-mediated degradation of A53T α-synuclein-EGFP in HEK293 cells was observed. (g) Omi does not cleave α-synuclein directly. Bacterially expressed GST-tagged A53T α-synuclein was incubated with HIS-tagged mature Omi in a protease buffer at 37°C for 30 min. β-Casein was used as a positive control to represent the protease activity of Omi. After incubation, samples were subjected to SDS-PAGE and visualized using Coomassie brilliant blue staining. (h) Increase of A53T α-synuclein-EGFP in a HeLa A53T α-synuclein-EGFP stable cell line by knockdown of Omi. (i) Quantitative data from (h) are shown. The data represent mean±S.E.M., n=3, *P<0.05, one-way ANOVA. GAPDH served as a loading control. (j) Increased tHtt-Q60-EGFP and tHtt-Q150-EGFP aggregation in Omi knockdown cells. HeLa cells with aggregates formed by tHtt-Q60-EGFP or tHtt-Q150-EGFP were counted. The data are shown as mean±S.E.M., n=3, *P<0.05, one-way ANOVA
To exclude the possibility that clearance of these disease proteins is due to the impact of co-transfection, we generated a mouse neuroblastoma (N2a) cell line stably expressing A53T α-synuclein-EGFP and examined the influence of Omi on A53T α-synuclein-EGFP clearance. A53T α-synuclein-EGFP levels were clearly decreased upon overexpression of Omi (Figure 3c and d). Similar results were obtained using a HeLa cell line stably expressing A53T α-synuclein-EGFP (Supplementary Figure S3b). However, in ATG5 KO MEF cells, Omi failed to induce A53T α-synuclein degradation (Figure 3e), suggesting that Omi-mediated A53T α-synuclein degradation depends on an autophagy/lysosome system. p62/Sequestosome 1, an endogenous autophagy substrate, is widely used to monitor the activation of autophagy.17 The amount of endogenous p62 protein was found to be lower in Omi-expressing cells than in control cells (Supplementary Figure S3c), indicating that Omi indeed activates the autophagic degradation pathway. To determine whether the proteasome system is also involved, we examined the effects of the proteasome inhibitor MG132 on A53T α-synuclein degradation induced by Omi. MG132 did not affect Omi-mediated A53T α-synuclein degradation (Figure 3f).
To exclude the possibility that the decrease in A53T α-synuclein was caused by its direct cleavage by Omi, we performed in vitro cleavage assays. A53T α-synuclein was not cleaved by Omi, whereas β-casein, the positive control, was greatly digested even at a low concentration of Omi (Figure 3g).
We then examined the effects of silencing Omi on the degradation of aggregation-prone proteins. Knockdown of Omi increased the A53T α-synuclein level in a HeLa cell line stably expressing A53T α-synuclein-EGFP (Figure 3h and i). In addition, the amount of the endogenous autophagy substrate p62 was increased in Omi knockdown cells compared with negative control cells (Supplementary Figure S3d). It was previously reported that the inhibition of autophagy increases aggregate formation in cell with mutant polyQ expression.19 In HeLa cells expressing tHtt-Q60 and tHtt-Q150, we observed that the percentage of cells with polyQ aggregates increased significantly when Omi was knocked down (Figure 3j). Taken together, these data suggest that Omi regulates the degradation of neurodegenerative disease proteins through autophagy.
Involvement of Beclin-1 and Hax-1 in Omi-induced autophagy
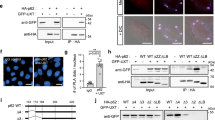
Wortmannin inhibits autophagy through the vps34 (class III PI3k)/Beclin-1 pathway, which is negatively controlled by antiapoptotic Bcl-2 family proteins.22 We therefore tested whether this pathway is involved in Omi-induced autophagy. We found that overexpression of Bcl-xL significantly attenuated Omi-induced autophagy (Figure 4a). Meanwhile, in Beclin-1-knockdown cells, Omi failed to enhance autophagy (Figure 4b). In HeLa cells overexpressing Beclin-1, knockdown of Omi also failed to decrease the basal level of autophagy (Figure 4c). These data suggest that Omi regulates autophagy in a Beclin-1-mediated pathway and functions upstream of Beclin-1. However, GST pull-down assays (Figure 4d) and immunoprecipitation assays (data not shown) showed that Omi does not bind to Beclin-1. Although in vitro purified Bcl-xL could bind Omi and its protease mutant (Supplementary Figure S4a), in vitro cleavage assays showed that Omi did not digest Bcl-xL (Figure 4e). Immunoprecipitation assays also showed that Omi did not interact with Bcl-xL (data not shown). These data suggest that Omi does not directly affect Bcl-xL to regulate autophagy.
Induction of autophagy by Omi in a Hax-1/Beclin-1-associated pathway. (a) Suppression of Omi-induced autophagy by Bcl-xL in HeLa cells. (b) Overexpression of Omi in Beclin-1-knockdown HeLa cells fails to activate autophagy. (c) Knockdown of Omi in Beclin-1-EGFP-overexpressing HeLa cells did not decrease the basal level of autophagy. (d) Omi does not preferentially bind to Beclin-1. In vitro purified His-Beclin-1 was incubated with GST-tagged Bcl-xL or mature Omi S276C or GST alone. After extensive washing with PBS, the His-Beclin-1 bound to GST-tagged proteins was observed by immunoblot using Beclin-1 antibodies. (e) Specific cleavage of Hax-1, but not Bcl-xL, by Omi. In vitro purified GST-tagged proteins were incubated with WT or protease-inactive S276C Omi for 60 min in a protease buffer at 37°C and were visualized by immunoblot using a GST antibody. (f) Reduction of Hax-1 by Omi. Increasing amounts of Omi-HA plasmids were transfected into HeLa cells along with Hax-1-EGFP plasmids. Cells were collected 48 h after transfection. (g) Decreased LC3-II levels in Hax-1-overexpressing HEK293 cells. (h) Inhibition of starvation-induced EGFP-LC3 puncta formation by Hax-1. HeLa cells were transfected with EGFP-LC3 alone with mock p3x-Flag-CMV vector or Flag-Hax-1 and were subjected to starvation (HBSS, 2 h) 48 h after transfection. (i) Quantitative analysis of (h). The data represent means±S.E.M., n=3, *P<0.05, one-way ANOVA. (j) Restoration of autophagy in Hax-1-overexpressing HeLa cells by Flag-Beclin-1. (k) Co-immunoprecipitation of Flag-Beclin-1 and endogenous Omi with Hax-1-EGFP in HEK293 cells. (l) Interaction of both WT Hax-1 and Δ-OBS-Hax-1 (Δ-Omi binding sequence Hax-1) with Beclin-1; the latter lost the ability to bind to Omi. (m) Decrease of WT, but not Δ-OBS-Hax-1, by Omi. (n) Δ-OBS-Hax-1 has a stronger ability to antagonize Omi-induced autophagy compared with WT Hax-1 in N2a cells. (o) Quantitative analysis of (n). The data represent means±S.E.M., n=3, *P<0.05, one-way ANOVA
Hax-1 is another Bcl-2 family-related antiapoptotic protein that was previously reported as a specific substrate of Omi.23 The cleavage of Hax-1 by Omi was confirmed, as shown in Figure 4e. In transfected HeLa cells, increased expression of Omi leads to decreased levels of Hax-1 (Figure 4f). Thus, we wondered whether Hax-1, as a specific substrate of Omi, is involved in autophagy regulation similarly to other Bcl-2 family proteins through interaction with and inactivation of Beclin-1. First, we examined whether Hax-1 could inhibit autophagy. In HEK293 cells, overexpression of Hax-1 decreased LC3-II levels (Figure 4g). In addition, Hax-1 reduced EGFP-LC3 puncta formation induced by starvation (Figure 4h and i), whereas knockdown of Hax-1 increased LC3 puncta formation (Supplementary Figure S4d and e). Furthermore, inhibition of autophagy by Hax-1 was partially rescued by overexpression of Beclin-1 (Figure 4j), whereas overexpression of Hax-1 attenuated Omi-induced autophagy (Supplementary Figure S4f). Interactions between Hax-1 and Beclin-1 were observed using immunoprecipitation assays (Figure 4k; Supplementary Figure S4g), GST pull-down assays (Supplementary Figure S4h) and immunofluorescent staining (Supplementary Figure S4i). It was previously reported that a deletion mutant of Omi (ΔOBS, Δ-Omi binding sequence, amino acids 201–213) could not bind to Omi.24 We therefore tested the effects of this mutant on autophagy. Hax-1-EGFP ΔOBS retained its ability to bind to Beclin-1, whereas it failed to interact with Omi (Figure 4l). Hax-1-EGFP ΔOBS was not digested by Omi (Figure 4m) but repressed Omi-induced autophagy more strongly than WT Hax-1 (Figure 4n and o); likely because it bypassed Omi to inhibit Beclin-1 function directly. These data collectively suggest that Omi may induce autophagy, at least in part, through digestion of the Beclin-1 inhibitory protein, Hax-1.
Decrease of constitutive autophagy in mnd2 mice
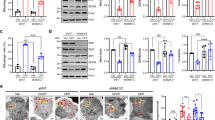
Motor neuron degeneration 2 (mnd2) mice harboring the Omi S276C mutant experienced severe neurodegeneration.7 We therefore examined whether the S276C mutation affects the ability of Omi to regulate autophagy in mnd2 mice. Embryonic fibroblast cells were collected from E14.5 mnd2 mouse embryos and age-matched WT mice, and the autophagic phenotype of these cells was examined. First, less activation of autophagy in response to tunicamycin and starvation was observed in MEF cells from mnd2 mice (Figure 5a), suggesting that the S276C mutation in Omi results in decreased autophagic response to various stresses. Meanwhile, more condensed nuclei were observed in mnd2 MEF cells treated with a higher dose of tunicamycin (Figure 5b and c), suggesting that the increased cell death induced by tunicamycin in mnd2 MEFs may result from insufficient autophagic induction (Supplementary Figure S5). Next, we examined the capability of MEF cells of mnd2 mice to degrade exogenous A53T α-synuclein. When equal amounts of plasmids were transfected into MEF cells from WT and mnd2 mice, we observed that the level of A53T α-synuclein-EGFP was significantly higher in MEF cells from mnd2 mice than in those from WT mice (Figure 5d), whereas the protein level of EGFP alone showed no difference. Last, we examined the protein levels in the brains of 35-day-old mnd2 mice. The LC3-II level was lower in mnd2 mice, although the protein levels of WT Omi in WT mice and S276C Omi in mnd2 mice were similar (Figure 5e and f). Both α-synuclein and p62 were dramatically increased, but DJ-1, another PD-associated protein, was not changed (Figure 5e and g). Furthermore, in the neurons of mnd2 mice, abnormal aggregates (dense spherical objects without a membrane) were frequently observed (Figure 5h). These data provide evidence that mnd2 mice, in which Omi is enzymatically inactivated, show decreased basal levels of autophagy and are also less capable of activating autophagy in response to stresses.
Decrease of constitutive autophagy by S276C mutant Omi in mnd2 mice. (a) Impaired autophagic activation in response to ER stress and starvation in mnd2 MEF cells. MEF cells from WT and mnd2 mice were treated with 1 μg/ml tunicamycin for 8 h or starved for 1 h. NT, no treatment. Primary cultured MEF cells from mnd2 or WT mice were isolated from E14.5 embryos. (b and c) Increased cell death observed in mnd2 MEF cells in response to prolonged ER stress. Wild-type or mnd2 MEFs were stimulated with 2 μg/ml tunicamycin for 24 h. The percentage of apoptotic cells was determined by Hoechst 33342 staining. The data are shown as means±S.E.M., *P<0.05, one-way ANOVA. (d) Reduction of the capability to degrade A53T α-synuclein in mnd2 MEF cells. MEF cells from WT or mnd2 mice were transfected with equal amounts of EGFP or A53T α-synuclein-EGFP plasmids and were subjected to immunoblot analysis. (e) Reduction of autophagy levels and increase in autophagic substrates in the mnd2 mouse brain. Brain lysates from 35-day-old WT mice and mnd2 mice were loaded for immunoblot analysis using the indicated antibodies. (f) Quantitative analysis of the relative density of LC3-II to LC3-I in mnd2 mouse brains compared with control brains. The data represent means±S.E.M., n=3, *P<0.05 one-way ANOVA. (g) Quantitative analysis of the relative density of α-synuclein with respect to that of GAPDH from (e). The data represent means±S.E.M., n=3, *P<0.05 one-way ANOVA. (h) Micrograph showing protein aggregates in central cortex neurons of 35-day-old mnd2 mice
Discussion
To the best of our knowledge, this is the first report to link Omi to autophagy. Bacterial HtrA proteins have key roles in the protein quality control system of prokaryotes, digesting accumulated proteins in the periplasmic space at higher temperatures.2 The eukaryote homologue of HtrA, Omi, is also activated under various stresses. Omi is reported to be upregulated by severalfold after heat shock or tunicamycin treatment.4 In our report, both mRNA and protein levels increased under tunicamycin treatment (Figure 2a; Supplementary Figure S2b). In a mouse model of kidney ischemia/reperfusion, protease activity of Omi obviously increased.25 These studies strongly suggest that Omi serves as a modulator of stress responses. Notably, the above stimuli that upregulate Omi also activate autophagy18, 26 (also shown in Figure 2a in our study), suggesting that Omi could be associated with autophagy. In this study, we provide further evidence that Omi directly regulates autophagy. Treatment of cells with ER stress inducers like tunicamycin leads to an activation of the unfolded protein response (UPR) and autophagy,18 as well as an increase in Omi levels27 (also shown in Figure 2). The autophagy system has a pivotal role in protecting cells against the death induced by various stressors.28 Recently, an important finding revealed that the loss of Omi activates the PERK branch of the UPR, with transcriptional upregulation of CHOP and modulation of a number of other genes leading to neuronal cell death.29 It is therefore possible that the upregulation of Omi and resulting activation of autophagy in response to stress are protective reactions, and the loss of this protective effect could result in sustained activation of the UPR. The upregulation of Omi by ER stress is similar to that observed for other prosurvival stress response proteins such as Grp78, which is also upregulated in response to ER stress; its knockdown also spontaneously activates UPR and upregulates CHOP.30
Omi is a key component needed to maintain mitochondrial integrity and the function of the respiratory chain, and loss of Omi produces a large number of damaged mitochondria.10, 29 We report here that Omi is responsible for the maintenance of constitutive autophagy (Figure 2). The autophagy system is a conserved pathway that eliminates damaged mitochondria.31 Loss of Omi function may concurrently suppress autophagy and yield damaged mitochondria. The inefficient elimination of malfunctioned mitochondria caused by suppression of autophagy may, in turn, increase the injury of cells. This finding is in agreement with previous findings that cells from mnd2 or Omi KO mice showed enhanced sensitivity to inducers that target the mitochondria.7, 10
In this report, we show that regulation of autophagy by Omi is tightly associated with Beclin-1 (Figure 4a–c). The Bcl-2 family protein Bcl-xL can block Omi-induced autophagy (Figure 4a). Interestingly, Hax-1, a Bcl-2 family-related protein and a specific substrate of Omi,23 binds to Beclin-1 and represses autophagy (Figure 4e–k). The effects of Hax-1 on autophagic inhibition can be partially rescued by overexpression of Beclin-1 (Figure 4l), and a deletion mutant of Hax that lacks the Omi binding sequence shows enhanced inhibition of Omi-induced autophagy (Figure 4l–o). It is therefore possible that Omi activates autophagy through interactions with or cleavage of Bcl-2 family proteins, such as Hax-1, that abolished their Beclin-1-antagonizing properties (Figure 6). Hax-1 is predominantly located in mitochondria.23 It has also been reported that Hax-1 can localize to the ER.32 Beclin-1 is located in both the mitochondria and the ER.22 In this study, Hax-1 co-localized with Beclin-1 (Supplementary Figure S4i). Because other Bcl-2 family proteins, such as Bcl-xl and Bcl-2, mainly reside in the mitochondria and ER and also bind tightly to Beclin-1,22, 33 we believe that the interaction between Hax-1 and Beclin-1 may occur in both the mitochondria and the ER. Hax-1 was first identified as a cleavage target of Omi in a yeast two-hybrid screen of Omi-interacting proteins. Degradation of Hax-1 by Omi was observed when cells were treated with various apoptotic inducers, such as cisplatin and H2O2.23 We report here that Hax-1, just like other Bcl-2 family-related proteins, is involved in autophagy through its interaction with and inhibition of Beclin-1. It is worth noting that Hax-1 functions not only as a substrate of Omi, but also as a regulator of the mitochondrial import of Omi through presentation of Omi to another mitochondrial protease, Parl.24 The loss of lymphocytes in response to certain stresses such as cytokine withdrawal was observed in both Hax-1-null mice and mnd2 mice.24 Cell death induced by ER (Figure 5) and mitochondrial stress7 was also enhanced in mnd2 cells. Collectively, these findings suggest that the interaction or balance between Hax-1 and Omi is crucial for various aspects of the cellular response. On the one hand, as an upstream regulator of Omi, Hax-1 ensures the proper localization of functional Omi to protect cells from stresses, on the other hand, as a specific substrate of Omi, Hax-1 can be digested by Omi to elicit autophagy in response to autophagic stimuli or to stimulate apoptosis23 when cells are ultimately sentenced to die. It will be interesting to explore how cells can maintain the equilibrium between Hax-1 and Omi under different circumstances.
Omi was recently reported to be associated with various types of neurodegeneration.34 Currently, increasing evidence has extensively associated autophagy with neurodegenerative diseases. Autophagy was first implicated in human neurodegenerative disease on the basis of the presence of autophagic vacuoles containing disease-related proteins in the brains of Alzheimer's disease (AD) patients.35 This finding was subsequently reported in Huntington's disease (HD) and other polyQ diseases, as well as in certain lysosomal storage disorders.36, 37 More persuasive evidence comes from autophagy-deficient animals, in which the conditional KO of ATG5 in the central nervous system results in neurodegeneration.15 A broad array of age-related neurodegenerative diseases is characterized by the accumulation and deposition of misfolded proteins in affected brains, including AD, PD and the polyQ diseases. Under these circumstances, autophagy-mediated degradation serves as a protein quality control system to safeguard cells from the activity of aberrant proteins and is therefore a protective response. In this report, we provide evidence that Omi enhances the degradation of the neurodegenerative disease proteins A53T α-synuclein and polyQ-expanded tHtt in an autophagy-dependent manner (Figure 3a–f). The knockdown of Omi decreases basal autophagy and increases A53T α-synuclein levels and the aggregate formation of tHtt-polyQs (Figure 3h–j). Constitutive and stress-induced autophagy, as well as degradation of the autophagy substrate A53T α-synuclein, was decreased in mnd2 MEF cells (Figure 5). Aggregate formation was observed in mnd2 neurons (Figure 5h). Increased α-synuclein and decreased autophagic activity were found in mnd2 brain lysates both before (data not shown) and after (Figure 5e) the onset of the disease. These data suggest that Omi-induced autophagy activity contribute in the degradation of proteins involved in neurodegenerative diseases. Correspondingly, it was reported that Omi protein levels were selectively decreased in striatal neurons in the brains of patients with HD, and overexpression of Omi protected cells against polyQ toxicity.38 Interestingly, a reduced Beclin-1 level was also observed in early AD.39 We therefore propose that Omi might be involved in the pathogenesis of neurodegenerative diseases through the modulation of the autophagy-mediated protein degradation pathway.
Materials and Methods
Animals and MEF cell isolation
Live heterozygotic mnd2 mice (HtrA2mnd2, C57BL/6J, stock number: 004608, 6 April 2007) were purchased from JAX (The Jackson Laboratory, Bar Harbor, ME, USA). Paired heterozygotic mice were used to breed homozygotic mnd2 mice. Mice were maintained in a specific pathogen-free environment. All animal experiments were approved by the animal welfare advisory committee of the University of Science and Technology of China. Adequate measures were taken to minimize pain and discomfort in compliance with national regulations. The offspring were born at a normal Mendelian ratio. Genotypes of offspring mice were identified through the protocol provided by JAX (http://jaxmice.jax.org/pub-cgi/protocols/protocols.sh?objtype=protocol&protocol_id=875). Primary MEF were established from E14.5 embryos as previously described10.
Cell culture, transfections and drug treatments
HeLa, HEK293, N2a and MEF cells were grown in Dulbecco's modified Eagle's medium (Gibco, Grand Island, NY, USA) containing 10% fetal calf serum with 100 μg/ml penicillin and 100 μg/ml streptomycin. ATG5 WT and KO MEF cells were kindly gifted by Dr. Noboru Mizushima. Transfections were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Stable cell lines were established using limiting dilutions with 400 μg/ml G418 (Gibco) and screened using immunoblotting or fluorescent microscopy. Rapamycin, 4′,6-diamidino-2-phenylindole (DAPI), Hoechst 33342, WM, tunicamycin and chloroquine were purchased from Sigma (St. Louis, MO, USA). MG132 was obtained from Calbiochem (San Diego, CA, USA).
Electron microscopy
Cells were fixed in suspension with 4% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.3), overnight at 4°C, and then post-fixed for 1 h at room temperature in 0.1 M Na-phosphate buffer (pH 7.4) containing 2% glutaraldehyde. After dehydration with graded ethanol and propylene oxide, we embedded cells in Epon. Thin sections were stained with uranyl acetate and lead citrate and then observed using a transmission electron microscope (JEOL-1230, Tokyo, Japan).
Plasmids
The EGFP-LC3 expression vector was kindly provided by Dr. Mizushima. The mammalian expression plasmid pcDNA3-Omi-Flag (encodes human Omi) was a kind gift from Dr. Hyangshuk Rhim. Full-length C-terminal MYC-tagged Omi was created by subcloning the PCR product amplified with primers 5′-AATGGTACCCCACCATGGCTGCGCCGAGGGCGG-3′ and 5′-CGCTCGAGTTCTGTGACCTCAGGGGTC-3′ into the pcDNA3.1 (+)/myc-HisA vector at KpnI and XhoI sites. Full-length C-terminal HA-tagged Omi was generated by excising full-length Omi cDNA from Omi-MYC and inserting it into the pKH3-HA vector at HindIII/XbaI sites. The mature form of Omi with a C-terminal MYC tag was created by subcloning the PCR product amplified with primers 5′-TAAAGATCTGATGGCCGTCCCTAGCCCGCCGCC-3′ and 5′-CGCTCGAGTTCTGTGACCTCAGGGGTC-3′ into the pcDNA3.1 (+)/myc-HisA vector at BglII and XhoI sites. The pET21a(+) vector expressing 6xHis-tagged mature Omi was created by subcloning the PCR product amplified with primers 5′-ATGTCGACAAGCCGTCCCTAGCCCGCCGCC-3′ and 5′-ATGCGGCCGCTTCTGTGACCTCAGGGGTCA-3′ into the pET21a(+) vector at SalI and NotI sites. The S276C mutant Omi-HA and HIS-Omi were created using the following primers: 5′-ATTGTTTGCTCTGCTCAGCGTCCA-3′ and 5′-GCCGGATGTGATCGTGTTCTGCAG-3′. The same sense mutations of WT and S276C Omi that were resistant to Omi siRNA 1 were created using a site-directed mutagenesis kit (Takara, Dalian, China) with primers 5′-GTAGTCGCAATGGGAAGTCCCTTTGCAC-3′ and 5′-GAATTCTCCTTGCCGGACATCAGCTGAG-3′. The same sense mutations of WT and S276C Omi that were resistant to Omi siRNA 2 were created using primers 5′-GGCTTCGTGGTGGCTGCC-3′ and 5′-CGAGCCGTTCGAGATAGG-3′. Full-length Hax-1 cDNA was amplified from a human fetal brain library (Invitrogen) using the primers 5′-GCGAATTCACCATGAGCCTCTTTGATCTCTTC-3′ and 5′-ATGTCGACATCCGGGACCGGAACCAACGTCCC-3′, and inserted into pEGFP-N1 (Clontech, Palo Alto, CA, USA), pGEX-5X-1 (Amersham Pharmacia Biotech, Arlington Heights, IL, USA) or p3x-Flag-CMV (Sigma) vectors at EcoRI and SalI sites. Full-length Beclin-1 cDNA was obtained using the primers 5′-GCTAAGCTTACCATGGAAGGGTCTAAGACGTC-3′ and 5′-GCCGTCGACTTTGTTATAAAATTGTGAGGACACC-3′, and was inserted into pEGFP-N3 (Clontech), pcDNA3.1-Flag (Invitrogen) or pCMV-myc (Sigma) vectors at HindIII and SalI (or XhoI) sites. Constructs expressing tHtt-60Q-EGFP, tHtt-150Q-EGFP and A53T-α-synuclein-EGFP were described previously.21, 40 The GST-A53T α-synuclein was generated by excising A53T α-synuclein cDNA from pEGFP-A53T α-synuclein at BglII/SalI sites and inserting it into pGEX-5X-1 at BamHI/XhoI sites. All constructs were sequenced to confirm their fidelity.
siRNAs
Typically, 35 pmol of each siRNA was transfected using Oligofectamine, according to the manufacturer's instructions (Invitrogen). Oligonucleotides were purchased from GenePharma (Shanghai, China) and had the following sequences:
- Si Omi 1 sense,:
-
5′-GGGGAGUUUGUUGUUGCCAdTdT-3′,
- Si Omi 1 antisense,:
-
5′-UGGCAACAACAAACUCCCCdTdT-3′;
- Si Omi 2 sense,:
-
5′-AACGGCUCAGGAUUCGUGGdTdT-3′,
- Si Omi 2 antisense,:
-
5′-CCACGAAUCCUGAGCCGUUdTdT-3′;
- Si Beclin-1 sense,:
-
5′-TGAGGATGACAGTGAACAGdTdT-3′,
- Si Beclin-1 antisense,:
-
5′-CTGTTCACTGTCATCCTCAdTdT-3′;
- Si Hax-1 sense,:
-
5′-AACCAGAGAGGACAAUGAUCUdTdT-3′,
- Si Hax-1 antisense,:
-
5′-AGAUCAUUGUCCUCUCUGGUUdTdT-3′;
- Si control sense,:
-
5′-UUCUCCGAACGUGUCACGUdTdT-3′,
- Si control antisense,:
-
5′-ACGUGACACGUUCGGAGAAdTdT-3′.
RT-PCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen) and then subjected to reverse transcription (RT)-PCR with the following primers: 5′-GACCTGACTGACTACCTC-3′ and 5′-GACAGCGAGGCCAGGATG-3′ for Actin and 5′-TAGGACCCCGGATCTCTGG-3′ and 5′-GACCCCAACCCCACAACAG-3′ for Omi.
Western blot analysis and antibodies
Cell extracts were lysed in 1 × RIPA lysis buffer (25 mM Tris-HCl (pH 7.6), 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate) in the presence of protease inhibitor cocktail (Roche, Indianapolis, IN, USA). Approximately 20 μg of cell lysates was separated on SDS-PAGE and transferred onto a PVDF membrane (Millipore, Bedford, MA, USA). Immunoblot analysis was carried out with the following primary antibodies: anti-α-synuclein (Cell Signaling Technology, Beverly, MA, USA), anti-Beclin-1 (Abcam, Cambridge, MA, USA), anti-DJ-1 (Chemicon, Temecula, CA, USA), anti-GAPDH (Chemicon), anti-ERK 1/2 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-GFP (Santa Cruz Biotechnology), anti-HA (Santa Cruz Biotechnology), anti-HtrA2 (R&D Systems, Minneapolis, MN, USA), anti-LC3 (Novas, Littleton, CO, USA), anti-MYC (Santa Cruz Biotechnology), anti-p62/SQSTM1 (Biomol, Exeter, UK) and anti-ubiquitin (Santa Cruz Biotechnology). The secondary antibodies, that is, sheep anti-mouse IgG-HRP antibodies or anti-rabbit IgG-HRP antibodies, were from Amersham Pharmacia Biotech. The proteins were visualized using an ECL detection kit (Amersham Pharmacia Biotech, Arlington Heights, IL, USA).
Statistical analysis
Western blot densitometry analysis of immunoblots from three independent experiments was performed using Photoshop 7.0 (Adobe). For EGFP-LC3 and immunofluorescence analysis, cells presenting a mostly diffuse distribution of EGFP-LC3 in the cytoplasm and nucleus were considered nonautophagic, whereas cells with >5 obvious intense punctate EGFP-LC3 aggregates with little or no nuclear localization were defined as autophagic. The data were analyzed using Origin 6.0 (Originlab, Northampton, MA, USA).
Immunoprecipitation
Cells transfected with the indicated plasmids were collected 48 h after transfection and lysed in TSPI buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM sodium chloride, 1 mM EDTA and 1% NP-40 supplemented with complete mini protease inhibitor cocktail (Roche). Cellular debris was removed by centrifugation at 12 000 × g for 30 min at 4°C. The supernatants were incubated with anti-GFP antibodies overnight at 4°C. After incubation, protein G Sepharose (Roche) was used for precipitation. The beads were washed with TSPI buffer four times and then eluted with SDS sample buffer for immunoblot analysis.
In vitro proteolytic cleavage assay
GST recombinant proteins or β-casein (Sigma) was incubated in the presence of 1–3 μg of 6 × HIS-tagged mature Omi in protease cleavage buffer (50 mM Tris-Hcl (pH 7.6), 1 mM dithiothreitol) for 45 min at 37°C. The reactions were stopped by the addition of 2 × SDS loading buffer and samples were boiled for 10 min at 95°C. Samples were then resolved by 12% SDS-PAGE and visualized by Coomassie brilliant blue staining.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde in PBS for 10 min at room temperature and then permeabilized in 0.5% Triton X-100 PBS for another 10 min. After preblocking with 5% fetal bovine serum for 1 h, cells were incubated with anti-HA or anti-MYC (Santa Cruz Biotechnology) antibody and then with rhodamine (red)- or FITC (green)-conjugated secondary antibody (Santa Cruz Biotechnology) for 2 h followed by incubation with DAPI (Sigma) for 5 min. Finally, the cells were observed using an inverted IX71 microscope system (Olympus, Tokyo, Japan).
Starvation
Cells were washed three times with PBS and incubated with 1 ml Hank's buffered salt solution (HBSS) without serum at 37°C.
Abbreviations
- KO:
-
knockout
- WT:
-
wild type
- MEF:
-
mouse embryonic fibroblast
- PD:
-
Parkinson's disease
- HtrA2:
-
high temperature requirement factor A2
- mnd2 :
-
motor neuron degeneration 2
- IAP:
-
inhibitor of apoptosis protein
- HD:
-
Huntington's disease
- htt:
-
huntingtin
- ATG:
-
autophagy-related gene
- LC3:
-
microtubule-associated protein 1 light chain 3
- polyQ:
-
polyglutamine
- ER:
-
endoplasmic reticulum
References
Pallen MJ, Wren BW . The HtrA family of serine proteases. Mol Microbiol 1997; 26: 209–221.
Clausen T, Southan C, Ehrmann M . The HtrA family of proteases: implications for protein composition and cell fate. Mol Cell 2002; 10: 443–455.
Suzuki Y, Takahashi-Niki K, Akagi T, Hashikawa T, Takahashi R . Mitochondrial protease Omi/HtrA2 enhances caspase activation through multiple pathways. Cell Death Differ 2004; 11: 208–216.
Gray CW, Ward RV, Karran E, Turconi S, Rowles A, Viglienghi D et al. Characterization of human HtrA2, a novel serine protease involved in the mammalian cellular stress response. Eur J Biochem 2000; 267: 5699–5710.
Martins LM, Turk BE, Cowling V, Borg A, Jarrell ET, Cantley LC et al. Binding specificity and regulation of the serine protease and PDZ domains of HtrA2/Omi. J Biol Chem 2003; 278: 49417–49427.
Suzuki Y, Imai Y, Nakayama H, Takahashi K, Takio K, Takahashi R . A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol Cell 2001; 8: 613–621.
Jones JM, Datta P, Srinivasula SM, Ji W, Gupta S, Zhang Z et al. Loss of Omi mitochondrial protease activity causes the neuromuscular disorder of mnd2 mutant mice. Nature 2003; 425: 721–727.
Strauss KM, Martins LM, Plun-Favreau H, Marx FP, Kautzmann S, Berg D et al. Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson's disease. Hum Mol Genet 2005; 14: 2099–2111.
Kawamoto Y, Kobayashi Y, Suzuki Y, Inoue H, Tomimoto H, Akiguchi I et al. Accumulation of HtrA2/Omi in neuronal and glial inclusions in brains with alpha-synucleinopathies. J Neuropathol Exp Neurol 2008; 67: 984–993.
Martins LM, Morrison A, Klupsch K, Fedele V, Moisoi N, Teismann P et al. Neuroprotective role of the Reaper-related serine protease HtrA2/Omi revealed by targeted deletion in mice. Mol Cell Biol 2004; 24: 9848–9862.
Gupta S, Singh R, Datta P, Zhang Z, Orr C, Lu Z et al. The C-terminal tail of presenilin regulates Omi/HtrA2 protease activity. J Biol Chem 2004; 279: 45844–45854.
Park HJ, Kim SS, Seong YM, Kim KH, Goo HG, Yoon EJ et al. Beta-amyloid precursor protein is a direct cleavage target of HtrA2 serine protease. Implications for the physiological function of HtrA2 in the mitochondria. J Biol Chem 2006; 281: 34277–34287.
Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S et al. Regulation of an ATG7-Beclin 1 program of autophagic cell death by caspase-8. Science 2004; 304: 1500–1502.
Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006; 441: 880–884.
Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006; 441: 885–889.
Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD et al. A protein conjugation system essential for autophagy. Nature 1998; 395: 395–398.
Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 2008; 4: 151–175.
Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 2006; 26: 9220–9231.
Ravikumar B, Duden R, Rubinsztein DC . Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet 2002; 11: 1107–1117.
Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC . Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem 2003; 278: 25009–25013.
Jana NR, Tanaka M, Wang G, Nukina N . Polyglutamine length-dependent interaction of Hsp40 and Hsp70 family chaperones with truncated N-terminal huntingtin: their role in suppression of aggregation and cellular toxicity. Hum Mol Genet 2000; 9: 2009–2018.
Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005; 122: 927–939.
Cilenti L, Soundarapandian MM, Kyriazis GA, Stratico V, Singh S, Gupta S et al. Regulation of HAX-1 anti-apoptotic protein by Omi/HtrA2 protease during cell death. J Biol Chem 2004; 279: 50295–50301.
Chao JR, Parganas E, Boyd K, Hong CY, Opferman JT, Ihle JN . Hax1-mediated processing of HtrA2 by Parl allows survival of lymphocytes and neurons. Nature 2008; 452: 98–102.
Faccio L, Fusco C, Chen A, Martinotti S, Bonventre JV, Zervos AS . Characterization of a novel human serine protease that has extensive homology to bacterial heat shock endoprotease HtrA and is regulated by kidney ischemia. J Biol Chem 2000; 275: 2581–2588.
Carloni S, Buonocore G, Balduini W . Protective role of autophagy in neonatal hypoxia-ischemia induced brain injury. Neurobiol Dis 2008; 32: 329–339.
Han C, Nam MK, Park HJ, Seong YM, Kang S, Rhim H . Tunicamycin-induced ER stress upregulates the expression of mitochondrial HtrA2 and promotes apoptosis through the cytosolic release of HtrA2. J Microbiol Biotechnol 2008; 18: 1197–1202.
He C, Klionsky DJ . Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet 2009; 43: 67–93.
Moisoi N, Klupsch K, Fedele V, East P, Sharma S, Renton A et al. Mitochondrial dysfunction triggered by loss of HtrA2 results in the activation of a brain-specific transcriptional stress response. Cell Death Differ 2009; 16: 449–464.
Li J, Ni M, Lee B, Barron E, Hinton DR, Lee AS . The unfolded protein response regulator GRP78/BiP is required for endoplasmic reticulum integrity and stress-induced autophagy in mammalian cells. Cell Death Differ 2008; 15: 1460–1471.
Kim I, Rodriguez-Enriquez S, Lemasters JJ . Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys 2007; 462: 245–253.
Sharp TV, Wang HW, Koumi A, Hollyman D, Endo Y, Ye H et al. K15 protein of Kaposi's sarcoma-associated herpesvirus is latently expressed and binds to HAX-1, a protein with antiapoptotic function. J Virol 2002; 76: 802–816.
Maiuri MC, Le Toumelin G, Criollo A, Rain JC, Gautier F, Juin P et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J 2007; 26: 2527–2539.
Vande Walle L, Lamkanfi M, Vandenabeele P . The mitochondrial serine protease HtrA2/Omi: an overview. Cell Death Differ 2008; 15: 453–460.
Dickson DW, Ksiezak-Reding H, Davies P, Yen SH . A monoclonal antibody that recognizes a phosphorylated epitope in Alzheimer neurofibrillary tangles, neurofilaments and tau proteins immunostains granulovacuolar degeneration. Acta Neuropathol 1987; 73: 254–258.
Jia K, Hart AC, Levine B . Autophagy genes protect against disease caused by polyglutamine expansion proteins in Caenorhabditis elegans. Autophagy 2007; 3: 21–25.
Massey AC, Zhang C, Cuervo AM . Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol 2006; 73: 205–235.
Inagaki R, Tagawa K, Qi ML, Enokido Y, Ito H, Tamura T et al. Omi/HtrA2 is relevant to the selective vulnerability of striatal neurons in Huntington's disease. Eur J Neurosci 2008; 28: 30–40.
Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA et al. The autophagy-related protein Beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest 2008; 118: 2190–2199.
Liu C, Fei E, Jia N, Wang H, Tao R, Iwata A et al. Assembly of lysine 63-linked ubiquitin conjugates by phosphorylated alpha-synuclein implies Lewy body biogenesis. J Biol Chem 2007; 282: 14558–14566.
Acknowledgements
We thank Dr. Noboru Mizushima (Tokyo Medical and Dental University, Japan) for the kind gifts of ATG5 WT and KO MEF cells and Dr. Hyangshuk Rhim (The Catholic University of Korea) for full-length Omi cDNA. We also thank Dr. Wen Hu (Anhui Provincial Hospital, China) for his support for TEM. This work was supported in part by the National Natural Sciences Foundation of China (nos. 30770664 and 30970921), the National High-tech Research and Development Program of China 973-projects (2006CB500703) and 863-projects (2006AA02A408) and the CAS Knowledge Innovation Project (KSCX2-YW-R-138).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by L Greene
Supplementary Information accompanies the paper on Cell Death and Differentiation website
Supplementary information
Rights and permissions
About this article
Cite this article
Li, B., Hu, Q., Wang, H. et al. Omi/HtrA2 is a positive regulator of autophagy that facilitates the degradation of mutant proteins involved in neurodegenerative diseases. Cell Death Differ 17, 1773–1784 (2010). https://doi.org/10.1038/cdd.2010.55
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cdd.2010.55
Keywords
This article is cited by
-
Omi inhibition ameliorates neuron apoptosis and neurological deficit after subarachnoid hemorrhage in rats
Genes & Genomics (2021)
-
Familial Parkinson’s Disease-Associated L166P Mutant DJ-1 is Cleaved by Mitochondrial Serine Protease Omi/HtrA2
Neuroscience Bulletin (2017)
-
Protease Omi facilitates neurite outgrowth in mouse neuroblastoma N2a cells by cleaving transcription factor E2F1
Acta Pharmacologica Sinica (2015)
-
Protease Omi cleaving Hax-1 protein contributes to OGD/R-induced mitochondrial damage in neuroblastoma N2a cells and cerebral injury in MCAO mice
Acta Pharmacologica Sinica (2015)
-
The protease Omi regulates mitochondrial biogenesis through the GSK3β/PGC-1α pathway
Cell Death & Disease (2014)