Abstract
To investigate the prevalence and diversity of Chlamydia spp. in domestic birds in China, oral and cloacal swabs of healthy chickens, ducks, geese and pigeons were collected nationwide from live-animal markets and examined by Chlamydia spp. 23 S rRNA gene FRET-PCR followed by high-resolution melting curve analysis and confirmatory sequencing. Overall, 26.2% of the birds (602/2,300) were positive for Chlamydia spp. and five Chlamydia spp. were identified. While occasional detection of C. suis and C. muridarum in poultry is reported here for the first time, the predominant chlamydial agent was C. gallinacea representing 63.8% of all positives (384/602) and 81.2% of positive chickens (359/442). Analysis of the C. gallinacea ompA phylogeny revealed at least 13 well segregated variants (serovars). Seven-month monitoring of C. gallinacea-infected chickens indicated that the infection was persistent. C. gallinacea-infected chickens remained without overt clinical disease, but showed body weight gains significantly reduced by 6.5–11.4% beginning in week 3 post-infection. This study indicates that C. gallinacea is the endemic chlamydial species in chickens, whereas C. psittaci dominates only in pigeons. Further studies are required to address the specific conditions under which C. gallinacea could act as an avian pathogen and possibly also a zoonotic agent.
Similar content being viewed by others
Introduction
The obligate intracellular bacteria, Chlamydia spp., are the etiological agents of chlamydiosis in wild and domestic birds, mammals and humans1,2,3,4,5. The family Chlamydiaceae currently contains the single genus Chlamydia (C.), which includes 11 recognized species, namely C. trachomatis, C. suis, C. muridarum, C. pneumoniae, C. abortus, C. caviae, C. felis, C. pecorum, C. psittaci and two recently introduced species, C. avium and C. gallinacea6.
Until recently, C. psittaci was considered to be the only pathogenic chlamydial species in birds, but recent evidence suggested that avian chlamydiosis may also involve C. gallinacea and C. avium7,8, C. abortus9, as well as C. pecorum or C. trachomatis10. Although most avian chlamydial infections remain asymptomatic11, they can lead to respiratory, enteric and ocular disease under appropriate conditions12,13.
So far, C. gallinacea has been isolated from chickens, ducks, guinea fowl, turkey and other domestic poultry in four European countries7,14. This new emerging agent was predominantly found in asymptomatic poultry, but a case of slaughterhouse workers presenting with atypical pneumonia was also reported in association with exposure to C. gallinacea-carrying chickens1. Both pathogenicity and possible zoonotic potential of C. gallinacea have yet to be systematically investigated.
As currently available data on chlamydial infections of domestic birds in China are confined only to three recent reports on C. psittaci in poultry15,16,17, this study was undertaken to explore the occurrence of individual chlamydial species in domestic birds, as well as to address the pathogenicity of C. gallinacea.
Results
Establishment of a FRET-qPCR platform to enable rapid and specific diagnosis for Chlamydia spp
The 23S rRNA gene-based FRET-qPCR established in this study detects all 11 Chlamydia species with high sensitivity and does not give a signal with other related bacteria. Multiple PCRs at 1 copy of chlamydial target per reaction yielded a Poisson distribution of positive and negative amplification reactions, indicating a robust PCR methodology that reliably amplified single target copies. Due to mismatch differences between the probes and the 23S rRNA sequences of 11 Chlamydia species (Fig. 1), melting curve analysis of the PCR products enabled us to classify the 11 Chlamydia species into 8 distinct groups (Fig. 2). For each Chlamydia species, the peaks and shapes of the melting curves were consistent at each of the four concentrations of the targets studied (10,000, 1,000, 100 and 10 copies of 23S rRNA/ 20 μl reaction system) (Fig. 2). A selection of 193 highly Chlamydia-positive samples based on FRET-qPCR (above 100 Chlamydia genomes/reaction, equivalent to 2,000 genomes/swab) were further verified in confirmatory PCRs targeting highly variable regions of the 16 S rRNA (697 bp) and 23S rRNA (329 bp) genes and by sequence determination of the amplification products for precise species identification (Table S1).
Alignment of primers and probes for 11 Chlamydia species.
Nucleotide sequences of the primers and probes (two upstream primers, one downstream primer, two fluorescein probes and one LCRed-640 probe) are shown above the corresponding nucleotide sequences of 11 Chlamydia species. The LCRed-640 probe and downstream primer show 0 or 1 mismatches with all 11 Chlamydia species. Upstream primer-1 and fluorescein probe-1 are specific for C. trachomatis, C. suis and C. muridarum and mismatches of chlamydial species are indicated by blue or black font. Upstream primer-2 and fluorescein probe-2 are specific for the remaining 8 Chlamydia species and mismatches are indicated by red or black font. The upstream primer and probes were used as shown while the downstream primer was used as reverse complement. Dots indicate that nucleotides are identical to those of primers/probes and dashes indicate deletion of nucleotides.
Tm differentiation of 11 Chlamydia species into eight groups through HRM genotyping analysis.
Following the completion of PCR, the Tm of probe hybridization to the targets was determined by high-resolution melting (HRM) curve analysis as the peak of the second derivative of the fluorescence released during a temperature increase from 38–85 °C. Based on the unique Tm distributions, 11 Chlamydia species are differentiated into 8 distinct groups: 50.6 °C for C. avium and C. caviae; 53.8 °C for C. pneumoniae; 54.9 °C for C. gallinacea; dual peaks of 55.8 °C and 61.4 °C for C. suis, 57.6 °C for C. felis, C. abortus and C. psittaci; a flat peak of 61.0 °C for C. trachomatis; 63.3 °C for C. pecorum; a flat peak of 65.0 °C for C. muridarum. For each Chlamydia species, five concentrations of the targets were used (only 100 and 10 copies of the gene/20 μl reaction system are shown here) and peaks and curve shape were consistent at all dilutions.
PCR prevalence of Chlamydia spp. in poultry
The Chlamydia FRET-PCR established in this study showed that 26.2% (602/2,300) of apparently healthy chickens, ducks, geese and pigeons from live-animal markets in 24 provinces of China were positive for Chlamydia spp. Among 471 Chlamydia-positive oral swabs, 372 (79.0%) contained 1–2,000 genomes and 99 swabs had higher loads up to 447,141 genomes/swab. Concerning cloacal swabs, 209 of 293 Chlamydia-positives (71.3%) had 1–2,000 genomes/swab and 84 swabs contained higher loads up to 966,977 genomes/swab. Overall, pigeons were most frequently infected (49.3%; 106/215), followed by chickens (24.7%; 442/1,791), ducks (24.0%; 43/179) and geese (9.6%; 11/115) (Table 1).
Considerable geographic differences in Chlamydia spp. prevalence were found. There was no Chlamydia spp. detection in five provinces, while PCR prevalence ranged from 1–26% in ten provinces and was above 30% in nine other provinces (Fig. 3). Overall, the positivity rate of oral swabs (22.0%; 471/2,138) was significantly higher (P < 10−4; Chi-square test) than that of cloacal swabs (16.1%; 293/1,824). In birds from which both oral and cloacal swabs were collected (n = 1,662), 162 were positive in both samples while 185 were only positive in oral swabs and 107 only in cloacal swabs. Of the 162 double-positive birds, 142 harbored the same Chlamydia species in both swabs.
Chlamydia spp. prevalence in 24 provinces of China.
Overall, 26.2% (602/2,300) of apparently healthy poultry from the live-poultry markets in 24 provinces of China were positive for Chlamydia spp. While Chlamydia spp. were not detected in 5 provinces (green: Shanxi, Tibet, Jilin, Liaoning and Shanghai), the PCR prevalences were between 1–26% in 10 provinces (yellow: Henan, Guangxi, Hebei, Hunan, Zhejiang, Xinjiang, Shaanxi, Inner Mongolia, Hubei and Gansu) and above 30% in 9 provinces (pink: Sichuan, Yunnan, Guangdong, Shandong, Anhui, Jiangxi, Fujian, Hainan and Jiangsu). Grey color was used to show the provinces where specimens were not available for this study. The colors ( for chicken,
for chicken,  for duck,
for duck,  for goose,
for goose,  for pigeon) and positions of filled circles indicate different poultry species and the sampling cities. This map was created by YY and CW using Adobe Illustrator CS5 (http://www.adobe.com/products/illustrator.html).
for pigeon) and positions of filled circles indicate different poultry species and the sampling cities. This map was created by YY and CW using Adobe Illustrator CS5 (http://www.adobe.com/products/illustrator.html).
Diversity of Chlamydia spp. in poultry
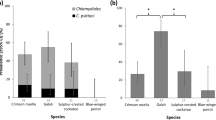
The use of FRET-qPCR followed by high-resolution melting curve analysis together with confirmatory PCRs targeting the variable domains of 16S rRNA and 23S rRNA genes enabled us to identify the presence of five Chlamydia spp. in the birds examined. The most common species was C. gallinacea (61.7%; 384/622), followed by C. psittaci (24.0%; 149/622), C. suis (10.5%; 65/622), C. muridarum (3.4%; 21/622) and C. pecorum (0.5%; 3/622) (Table 1). C. gallinacea and C. psittaci were the only ones present in all four avian species investigated. The former was detected in 81.2% of the Chlamydia-positive chickens (359/442). In contrast, 94.3% of the Chlamydia-positive pigeons (100/106) carried C. psittaci (Table 1).
Comparative phylogenetic analysis demonstrated that the five Chlamydia species identified in this study were all closely related to their respective Chlamydia spp. sequences, based on both 16S rRNA (0, 1, 3 mismatches/697 nucleotides; 99.6–100% similarity) (Fig. 4, Table S2) and 23S rRNA genes (0–3 mismatches/329 nucleotides; 99.1–100% similarity) (Fig. 4, Table S3).
Phylogeny of Chlamydia species and strains identified in this study.
A 697 bp variable region of the 16S rRNA gene is shown in the left panel and a 329 bp variable region of the 23S rRNA gene in the right panel. All 11 Chlamydia spp. type strains are shown in black font (Chlamydia sp, name of strain and sequence accession number) and the Chlamydia spp. identified in this study are shown in red font. Branch lengths are measured in nucleotide substitutions and numbers show branching percentages in bootstrap replicates. Scale bar shows the percentage sequence diversity.
Isolation of C. gallinacea strains and ompA polymorphism
Four C. gallinacea isolates (JXC1-4) were obtained from an oral swab and three cloacal swabs of chickens from Jiangxi province by propagation in Hep-2 cells. Their ompA sequences (GenBank accession number: KT692977) were identical to each other and showed 85.7% similarity to the only complete ompA sequence available in GenBank (strain 08-1274/3; accession number: AWUS01000004). Phylogenetic comparison showed that the ompA gene of the C. gallinacea strains is highly polymorphic and at least 13 new variants that were deeply separated and based on the sequence diversity clearly represent serovars18, were identified in addition to the published ompA sequences (Fig. 5, Figs S1 and S2).
Phylogeny of ompA variable domains 1–2 and VD 3–4.
A 421 bp region encompassing VD 1–2 is shown on the left panel and a 435 bp region encompassing VD 3–4 on the right panel. European C. gallinacea sequences deposited in GenBank are shown in black font (name of strain and accession number) and strains identified in this study in red font. The inability to amplify every target from every swab specimen resulted in strain differences between the phylograms. Branch lengths are measured in nucleotide substitutions and numbers show branching percentages in bootstrap replicates. Scale bar represents the percentage sequence diversity.
Persistent infection of C. gallinacea in chickens
Monitoring for Chlamydia spp. in naturally-infected chickens in the course of seven months demonstrated persistent C. gallinacea infection (Fig. 6), whereas infections with other chlamydial species were observed only transiently. Thirty-one free-range chickens from a mountain village were naturally infected with C. suis (28/31; 90.3%), C. psittaci (2/31; 6.5%) and C. gallinacea (1/31; 3.2%). As shown in Fig. 6, three weeks after transferring these 31 chickens to a containment animal facility, C. suis and C. psittaci disappeared, while the C. gallinacea carrier status was maintained (93.5%; 29/31) until the end of monitoring 3 months later.
A seven-month monitoring of Chlamydia spp. in naturally infected chickens moved from mountain village to animal facility.
FRET-qPCR was used to detect DNAs of Chlamydia spp. in oral and cloacal swabs of chickens which were moved from a mountain village in Jiangxi province (first three samplings) to a containment animal facility in Jiangsu province (last six samplings). Of the chickens tested in the mountain village, 74.4% (35/47) and 40.9% (36/88) were positive for Chlamydia spp. Thirty-one randomly-selected Chlamydia spp.-positive chickens (third sampling) were moved to the animal facility on November 27, 2014. Interestingly, four Chlamydia species were detected in the first three samplings when the chickens were in mountain village and the most prevalent species was C. suis [31/35 (88.6%), 30/36 (83.3%), 28/31 (90.3%)]. In the first two sampling time points in the animal facility, chlamydial positivity declined precipitously. In the final 4 sampling time points C. suis and C. psittaci had disappeared from the chickens, but C. gallinacea had completely overtaken as the only chlamydial species detected. While no signs of disease were observed, C. gallinacea-positivity became very high during the time (29/31, 30/30, 19/23 and 19/21).
In the five euthanized chickens of this group, chlamydial DNA was detected in all cloacal samples, in lung, heart and oral swab (n = 4), liver, trachea, kidney and pancreas (n = 3) and spleen and whole blood (n = 2). The average bacterial genome numbers were highest in cloacal swabs (105.15±0.97 [SEM]/swab), followed by oral swabs (103.80±2.62/swab), lung (103.38±1.99/gram), heart (103.35±1.89/gram), kidney (103.07±2.96/gram), liver (102.82±2.61/gram), pancreas (102.61±2.45/gram), trachea (102.51±2.30/gram), spleen (102.00±2.74/gram) and whole blood (101.73±2.45/ml).
C. gallinacea inoculation of chicken embryos and broiler chickens
To investigate the clinical course and economic impact of C. gallinacea infections in chickens, we performed preliminary infection experiments in chicken embryos and chickens. Yolk sac inoculation of one week-old chicken embryos (n = 5) with C. gallinacea resulted in 80% mortality of the embryos 8–10 days later (day 8, n = 2; day 9, n = 1; day 10, n = 1). FRET-qPCR determined the highest C. gallinacea loads in the yolk sac membrane (2 × 107 genomes/mg), followed by yolk (4 × 106 genomes/mg) and allantoic fluids (2 × 106 genomes/mg). All control chicken embryos (n = 5) inoculated with sucrose-phosphate-glutamate (SPG) buffer only matured normally and hatched as healthy chickens.
No signs of clinical disease were observed in C. gallinacea-inoculated broiler chickens and mock-inoculated chickens. Body weights did not differ significantly between C. gallinacea- and mock-inoculated chickens in the first two weeks pi. However, subsequently in weeks 3–5 pi, C. gallinacea-inoculated chickens showed significantly lower body weights than the mock-inoculated chickens. Overall, C. gallinacea-inoculated chickens showed 8.2%, 11.4% and 6.5% lower weekly weight gains as compared to the control group in weeks 3–5 pi (Fig. 7).
C. gallinacea reduces body weight of chickens by up to 11.4%.
Body weight (±SEM, left ordinate, in black) is shown for C. gallinacea-inoculated AA broiler chickens (filled circle) and mock-inoculated chickens (open circle). The difference in weekly body weight gain is shown in the right ordinate (in red). Compared to mock-inoculated chickens, C. gallinacea-inoculated chickens had significantly lower body weights (883 ± 54 [SEM] vs. 962 ± 61 g; 1165 ± 80 vs. 1315 ± 93 g; 1539 ± 120 g vs. 1645 ± 133 g) and lower body weight increase (8.2%, 11.4%, 6.5%, p < 0.0009) at 3, 4 and 5 weeks post inoculation.
Discussion
There are 11 closely related and distinct Chlamydia species and most animals are susceptible to multiple Chlamydia species, resulting in often asymptomatic infections with low bacterial burdens. While species-specific PCRs have been successfully used to detect individual Chlamydia species such as C. pneumoniae19, C. trachomatis20,21, C. abortus22, C. felis23, C. avium24, C. gallinacea14, C. psittaci and C. pecorum25 and C. psittaci and C. abortus26, there is a substantial need to establish a genus-specific FRET-qPCR that simultaneously in a single step detects single target copies of all Chlamydia spp. and differentiates the amplification product(s) by species. In this study, a novel FRET-qPCR followed immediately by high-resolution melting analysis of the reaction enables the sensitive detection of all chlamydial species and simultaneous differentiation of 11 species into 8 groups. Discriminatory PCRs for sequencing can be performed on Chlamydia-positive samples if further differentiation between Chlamydia spp. with similar Tm (between C. avium and C. caviae; or between C. felis, C. abortus and C. psittaci) is necessary (Fig. 2). This PCR methodology provides a practical and convenient tool for the epidemiological investigation of Chlamydia spp. in a variety of sources.
C. psittaci has long been considered the main chlamydial species in poultry. While C. psittaci was detected in all four avian species investigated in this study, our data showed that C. psittaci was the dominant species only in pigeons, representing 94.3% (100/106) of Chlamydia-positive pigeons but much lower percentages (9.1–16.3%) in chickens, ducks, or geese (Table 1). The positivity of Chlamydia spp. in this study is considerably higher than in other reports. For example, the overall prevalence for C. psittaci was 7.9% (26/331) of fecal samples collected from feral pigeons in the Netherlands27 and Tanaka et al. reported detection of C. psittaci in 22.2% (103/463) of feral pigeons in Japan28. C. psittaci was detected in 18 out of 19 Belgian chicken farms by culture and PCR29. Furthermore, it was reported that individuals working in a poultry slaughterhouse with C. psittaci-positive ducks presented with cough or flu-like symptoms30. Since zoonotic C. psittaci was detected in healthy market birds, health workers and consumers should be aware of the possibility of contracting C. psittaci infection from such birds, particularly from pigeons.
The results of this study are consistent with the current idea that the epidemiology of avian chlamydiosis is complex and Chlamydia infections in birds cannot be automatically ascribed to C. psittaci. In agreement with the report from Sachse et al.10, C. pecorum was also detected in chickens in this study. Earlier studies also demonstrated the presence of C. abortus, C. avium and C. trachomatis in birds8,9,10, whereas these species have not been identified in this study. However, we reported for the first time the detection of C. suis and C. muridarum in chickens, ducks and pigeons. With the exception of the dominance of C. psittaci in pigeons, these chlamydial species represented overall sporadic detections, typically confined to “hotspots”. Transmission of these chlamydiae probably occurred when the avian species had close contact to the natural hosts of C. pecorum, C. suis, or C. muridarum.
In contrast, our large and China-wide study involving 2,300 domestic birds demonstrates unambiguously that C. gallinacea is the endemic chlamydial species in chickens. The agent was detected in 81.2% of the Chlamydia-positive chickens in all four bird species and in 18 of 24 Chinese provinces investigated in this study (Table 1). Phylogenetic analysis of the variable domains of the OmpA protein of C. gallinacea identified at least 13 well segregated genetic variants (probably serovars), with 10 of them clearly different from known European isolates. This high diversity is consistent with immunoselection of ompA variants in endemic infection conditions31,32. For the time being, poultry must be considered the main natural host for C. gallinacea. However, as observed in this study, it is not unusual that Chlamydia spp. cross host barriers and infect new hosts or even humans. This may explain that slaughterhouse workers showed atypical pneumoniae after being exposed to C. gallinacea-carrying chickens1.
The epidemiological survey of Chlamydia spp. enabled us to identify a flock of Chlamydia-positive chickens in a mountain village that asymptomatically carried C. suis and, to a lesser extent, C. gallinacea, C. psittaci and C. muridarum. Interestingly, three weeks after transfer of 31 birds from that flock to a containment animal facility, C. suis and C. psittaci completely disappeared while C. gallinacea became the most prevalent species. The latter was also detected in blood, oral and cloacal swabs and 8 other organs. This persistent infection with C. gallinacea in chickens represents a major epidemiological reservoir. Previous carriage of C. suis and C. muridarum in chickens may have been the result of transient infection from continuous contact to pigs and wild mice in the mountain village.
As evident in the chicken flock from the mountain village, chickens carrying C. gallinacea usually do not show clinical signs. To characterize its pathogenic potential, we utilized a C. gallinacea isolate obtained in this study to inoculate chicken embryos and SPF broiler chickens and followed the course of infection. While inoculation of chicken embryos resulted in high mortality, intranasal inoculation of 7-day-old broiler chickens with C. gallinacea did not generate any clinical signs. However, the infected chickens showed significantly lower body weight gain from 3 weeks pi onwards. This is consistent with the report that asymptomatic endemic C. pecorum infections reduce growth rates in calves by up to 48 percent33,34. In combination with the ubiquitous endemic nature of C. gallinacea infection in chickens, this growth-suppressing effect of 10% or more may well be of major economic significance.
In conclusion, by using FRET-qPCR, we have demonstrated that Chlamydia spp. are common in healthy domestic birds traded on local live-animal markets in China. Our data confirmed that C. psittaci is the dominant species in pigeons, while the new emerging C. gallinacea is the endemic species in chickens. This investigation supported the notion that C. gallinacea is not a commensal, but a pathogen of moderate pathogenicity and that persistent infection leads to reduced body weight gain of broilers. It is imperative to further investigate the agent's pathogenicity, host spectrum, transmission mechanisms and possible zoonotic potential.
Materials and Methods
Oral and cloacal swabs
Between December 2013 and February 2014, a total of 3,962 oral and cloacal swabs were collected from 2,300 apparently healthy poultry (1,791 chickens, 215 pigeons, 179 ducks and 115 geese) in live-poultry markets in 24 provinces that encompass all seven geographical regions of China. Sterile cotton swabs were inserted into the throat and cloaca of the chicken and then turned slowly to absorb the fluid sample. All swabs were immediately placed in sterile tubes containing 400 μl DNA/RNA stabilization buffer (Roche Molecular Biochemicals, Indianapolis, IN, USA) and then stored at −80 °C until DNA was extracted as described below. Most of the poultry were free-range raised on family farms and few animals were originated from industrial poultry operations. Oral and cloacal swabs were obtained from each bird (n =1,662), but in some cases only oral (n = 476) or cloacal swabs (n = 162) were collected. All work in this study was reviewed and approved by the Institutional Animal Care and Use Committee of the Yangzhou University College of Veterinary Medicine (YZUCVM-IACUC 2013#87). The experiments were performed in accordance with the approved IACUC protocols.
DNA extraction
The High-Pure PCR Template Preparation Kit (Roche Molecular Biochemicals, Indianapolis, IN, USA) was used to extract total nucleic acids from oral and cloacal swabs, the whole blood and organs and tissues from chickens, according to the manufacturer’s instructions and described before35. The extracted DNA was eluted in 200 μl elution buffer.
FRET-qPCR for detection of Chlamydia spp
Primers and probes
The 23S rRNA sequences for all 11 Chlamydia species were obtained from GenBank: C. pneumoniae (NR076161, U76711), C. trachomatis (NR103960, NR076160), C. psittaci (NR102574, JN426968), C. abortus (NR077001, U76710), C. pecorum (NR103180, U68434), C. muridarum (NR076163, U68436), C. suis (U68420, AF481047), C. felis (NR076260, AP006861), C. caviae (NR076195, AE015925), C. avium (NR121988) and C. gallinacea (AWUS01000004). The sequences were aligned using Clustal Multiple Alignment (Vector NTI, Invitrogen, USA) to identify conserved and variable regions suitable for primers and probes that could detect all 11 Chlamydia species and largely differentiate the Chlamydia species by differential Tm. The master mix of the PCR contained two upstream primers, one downstream primer, two fluorescein probes and one LCRed 640 probe (Fig. 1, Table S1). This PCR amplified a 168 bp fragment of the Chlamydia spp. 23S rRNA gene and followed the design described by DeGraves et al.25, but added an upstream primer and a fluorescein probe.
Thermal cycling and melting curve analysis
The FRET-qPCR for Chlamydia spp. was performed in a LightCycler 480-II real-time PCR platform using a high-stringency 18-cycle step-down temperature protocol without fluorescence acquisition followed by 30 fluorescence acquisition cycles with a hybridization temperature of 51 °C25. The PCR master mix contained two upstream primers (0.5 μM), one downstream primer (1.0 μM) and three probes (0.2 μM). High-resolution melting curve analysis was performed following the completion of the PCR36 and data were analyzed as 640 nm: 530 nm (F4/F1) fluorescence ratios with the first derivative of F4/F1 (-d (F4/F1)/dt) evaluated. The fluorescent signals and melting peaks for positive controls, negative controls and other related bacteria were read to determine their positivity.
Specificity
To test the specificity of the FRET-PCR established in this study, the DNA of three Chlamydia species (C. pneumoniae strain CDC/CWL-029, ATCC VR-1310; C. trachomatis serovar D strain UW-3/Cx, ATCC VR-885; C. psittaci strain B577, ATCC VR-656) and 8 plasmids manufactured with the pUC57 cloning vector (GenScript, Nanjing, Jiangsu, China) containing an appropriate portion of the 23 S rRNA gene of the remaining 8 Chlamydia spp. were used as positive controls. As negative controls, we used DNAs extracted from related bacteria, including Neisseria sicca, Riemerella anatipestifer, Escherichia coli, Salmonella Enteritidis, Staphylococcus aureus and Streptococcus pyogenes (kindly provided by the Yangzhou University College of Veterinary Medicine). The specificity of the PCR was further verified by electrophoresis of PCR products through 2% agarose gels (BIOWEST®, Hong Kong, China), purification using the QIAquick Gel Purification Kit (Qiagen, Valencia, CA, USA) and sequencing with forward and reverse primers (GenScript, Jiangsu, Nanjing, China).
Sensitivity
To test the sensitivity of the FRET-qPCR, PCR amplification products from 11 Chlamydia species were gel purified with the QIAquick Gel Extraction Kit (Qiagen, Valencia, CA, USA) and quantified using the PicoGreen DNA fluorescence assay (Molecular Probes, Eugene, OR, USA). To obtain quantitative standards, the molarity of the Chlamydia DNA was determined using the calculated molecular mass of the PCR products, which then were adjusted to provide solutions containing 10,000, 1,000, 100, 10, 1 gene copies per PCR reaction in T10E0.1 buffer as described previously37.
Confirmatory sequencing
Chlamydia-positive samples based on FRET-qPCR and high-resolution melting curve analysis were further amplified for DNA sequencing to confirm the identified Chlamydia spp. PCRs were designed to target a variable region of 16S rRNA gene (697 bp) and a highly variable region of 23S rRNA gene (329 bp) for all 11 Chlamydia spp. (Table S1). To investigate the polymorphism of ompA gene in the C. gallinacea isolates and clinical specimens, the ompA PCR-1 was designed to amplify the complete ompA gene of C. gallinacea (1,188 bp) while ompA PCR-2 and ompA PCR-3 amplify the regions of variable domains (VD) 1–2 (435 bp) and VD 3–4 (421 bp) of C. gallinacea (Table S1). These PCRs were performed as described above for FRET-qPCR. The PCR products were electrophoresed through 2% agarose gel (BIOWEST®, Hong Kong, China) and purified for automated DNA sequencing (GenScript, Jiangsu, Nanjing, China).
Isolation of C. gallinacea
Human Epidermoid Carcinoma-2 cells (Hep-2 cells) were grown in Iscove’s Modified Dulbecco’s Medium (IMDM, Life Technology, USA) with 10% fetal bovine serum (Life Technology, USA) and amphotericin B (250 μg/ml, Amresco, USA) at 37 °C with 5% CO2 in a humidified cabinet for 24–48 h. Swabs in 200 μl SPG buffer from animals that were C. gallinacea-positive were transferred for chlamydial isolation into sterile tubes and vortexed with sterile magnetic beads for 3 min. After centrifugation at 1,250 g for 5 min, supernatants were transferred for incubation into 25 cm2 cell culture flasks with 7.6 ml IMDM and 0.4 ml antibiotics dissolved in IMDM (vancomycin 100 μg/ml, streptomycin 100 μg/ml, kanamycin 100 μg/ml, amphotericin B 3.75 μg/ml and gentamycin 10 μg/ml).
Monitoring of Chlamydia spp. naturally-infected chickens
Thirty-one locally-bred, free-range chickens, around 42 weeks of age, were transported from a mountain village in Jiangxi Province to a containment animal facility at the Poultry Institute in Jiangsu province. This mountain village has several swine farms and many free-range chickens. The chickens appeared healthy on arrival and were housed on the floor with free access to antibiotics-free food and water. Oral and cloacal swabs were collected aseptically and placed into tubes with 600 μl SPG buffer for isolation of Chlamydia spp. and for DNA extraction. Two months after being moved into the animal facility, five randomly-selected chickens were euthanized to test for Chlamydia DNA in their blood samples, oral and cloacal swabs and organs (tracheas, heart, liver, spleen, lung, kidney and pancreas) by FRET-qPCR.
Chicken embryos, broilers and C. gallinacea infection
White Leghorn chicken embryos purchased from Beijing Merial Vital Laboratory Animal Technology Co., Ltd (Beijing, China) were incubated with rotation at 37.8 °C and 60% relative humidity. The yolk sac was inoculated at embryonic development day 7 with 200 μl C. gallinacea suspension (2 × 106 genomes) or SPG buffer (as mock inoculation) as described38,39. Candling was performed once daily to determine the vitality of the chicken embryos.
One-day-old SPF AA broiler chickens obtained from Sandeli Animal Husbandry Development Co., Ltd (Zhenjiang, China) were individually tagged and housed in a containment level 2 facilities with free access to antibiotics-free food and water. After one week, the chickens were separated into two groups that were inoculated intranasally with 20 μl of 2 × 106 genomes of C. gallinacea diluted in SPG buffer (n = 15) or an equal volume of SPG buffer as control group (n = 15). The body weight of each chicken was recorded weekly before the inoculation and up to 5 weeks post inoculation while the oral and cloacal swabs were collected to detect the Chlamydia DNAs by FRET-qPCR.
Statistical analysis
Comparisons of the PCR prevalences of Chlamydia spp. in oral and cloacal swabs were analyzed by the Chi-Square Test. The two-tailed Tukey honest significant difference (HSD) test (Statistica, StatSoft, Tulsa, USA) was performed to compare the means of the body weight in mock- and C. gallinacea-inoculated chickens. Differences at P ≤ 0.05 were considered significant.
Additional Information
How to cite this article: Guo, W. et al. Chlamydia gallinacea, not C. psittaci, is the endemic chlamydial species in chicken (Gallus gallus). Sci. Rep. 6, 19638; doi: 10.1038/srep19638 (2016).
References
Laroucau, K. et al. Isolation of a new chlamydial agent from infected domestic poultry coincided with cases of atypical pneumonia among slaughterhouse workers in France. Infect. Genet. Evol. 9, 1240–1247 (2009).
Rohde, G., Straube, E., Essig, A., Reinhold, P. & Sachse, K. Chlamydial zoonoses. Dtsch. Ärztebl. Int. 107, 174–180 (2010).
Reinhold, P., Hartmann, H. & Constable, P. D. Characterization of acid-base abnormalities in pigs experimentally infected with Chlamydia suis. Vet. J. 184, 212–218 (2010).
Dean, D., Rothschild, J., Ruettger, A., Kandel, R. P. & Sachse, K. Zoonotic Chlamydiaceae species associated with trachoma, Nepal. Emerg. Infect. Dis. 19, 1948–1955 (2013).
Knittler, M. R. & Sachse, K. Chlamydia psittaci: update on an underestimated zoonotic agent. Pathog. Dis. 73, 1–15 (2015).
Sachse, K. et al. Emendation of the family Chlamydiaceae: proposal of a single genus, Chlamydia, to include all currently recognized species. Syst. Appl. Microbiol. 38, 99–103 (2015).
Sachse, K. et al. Evidence for the existence of two new members of the family Chlamydiaceae and proposal of Chlamydia avium sp. nov. and Chlamydia gallinacea sp. nov. Syst. Appl. Microbiol. 37, 79–88 (2014).
Sachse, K. & Laroucau, K. Avian chlamydiosis: two more bacterial players discovered. Vet. J. 200, 347–348 (2014).
Pantchev, A., Sting, R., Bauerfeind, R., Tyczka, J. & Sachse, K. New real-time PCR tests for species-specific detection of Chlamydophila psittaci and Chlamydophila abortus from tissue samples. Vet. J. 181, 145–150 (2009).
Sachse, K., Kuehlewind, S., Ruettger, A., Schubert, E. & Rohde, G. More than classical Chlamydia psittaci in urban pigeons. Vet. Microbiol. 157, 476–480 (2012).
Kaleta, EF. & Taday, E. M. A. Avian host range of Chlamydophila spp. based on isolation, antigen detection and serology. Avian Pathol. 32, 435–462 (2003).
Vanrompay, D., Ducatelle, R. & Haesebrouck, F. Chlamydia psittaci infections: a review with emphasis on avian chlamydiosis. Vet. Microbiol. 45, 93–119 (1995).
Knittler, M. R. et al. Chlamydia psittaci: new insights into genomic diversity, clinical pathology, host-pathogen interaction and antibacterial immunity. Int. J. Med. Microbiol. 304, 877–893 (2014).
Zocevic, A. et al. Molecular characterization of atypical Chlamydia and evidence of their dissemination in different European and Asian chicken flocks by specific real-time PCR. Environ. Microbiol. 14, 2212–2222 (2012).
Cong, W. et al. Seroprevalence of Chlamydia psittaci infection in market-sold adult chickens, ducks and pigeons in north-western China. J. Med. Microbiol. 62, 1211–1214 (2013).
Ling, Y. et al. Epidemiology of Chlamydia psittaci Infection in Racing Pigeons and Pigeon Fanciers in Beijing, China. Zoonoses Public Health. 62, 401–406 (2015).
Zhao, J. M., Rong, G., Zhou, H. L. & Hou, G. Y. Seroprevalence and risk factors of Chlamydia psittaci infection in domestic geese Anser domestica, in Hainan province of China. Acta. Trop. 145, 23–25 (2015).
Rahman, K. S. et al. Defining species-specific immunodominant B cell epitopes for molecular serology of Chlamydia species. Clin. Vaccine Immunol. 22, 539–552 (2015).
Hardick, J. et al. Real-time PCR for Chlamydia pneumoniae utilizing the Roche Lightcycler and a 16S rRNA gene target. J. Mol. Diagn. 6, 132–136 (2004).
Eickhoff, M. et al. Ultra-rapid detection of Chlamydia trachomatis by real-time PCR in the LightCycler using SYBR green technology or 5′-nuclease probes. Clin. Lab. 49, 217–225 (2003).
Solomon, A. W. et al. Strategies for control of trachoma: observational study with quantitative PCR. Lancet. 362, 198–204 (2003).
Creelan, J. L. & McCullough, S. J. Evaluation of strain-specific primer sequences from an abortifacient strain of ovine Chlamydophila abortus (Chlamydia psittaci) for the detection of EAE by PCR. Microbiol. Lett. 190, 103–108 (2000).
Helps, C., Reeves, N. & Tasker, S. & Harbour, D. Use of real-time quantitative PCR to detect Chlamydophila felis infection. J. Clin. Microbiol. 39, 2675–2676 (2001).
Zocevic, A. et al. A real-time PCR assay for the detection of atypical strains of Chlamydiaceae from pigeons. PLoS One. 8, e58741 (2013).
DeGraves, F. J., Gao, D., Hehnen, H. R., Schlapp, T. & Kaltenboeck, B. Quantitative detection of Chlamydia psittaci and C. pecorum by high-sensitivity real-time PCR reveals high prevalence of vaginal infection in cattle. J. Clin. Microbiol. 41, 1726–1729 (2003).
Opota, O. et al. Improving the molecular diagnosis of Chlamydia psittaci and Chlamydia abortus infection with a species-specific duplex real-time PCR. J. Med. Microbiol. 64, 1174–1185 (2015).
Heddema, E. R. et al. Prevalence of Chlamydophila psittaci in fecal droppings from pigeons in Amsterdam, The Netherlands. Appl. Environ. Microbiol. 72, 4423–4425 (2006).
Tanaka, C., Miyazawa, T., Watarai, M. & Ishiguro, N. Bacteriological survey of feces from feral pigeons in Japan. J. Vet. Med. Sci. 67, 951–953 (2005).
Lagae, S., Kalmar, I., Laroucau, K., Vorimore, F. & Vanrompay, D. Emerging Chlamydia psittaci infections in chickens and examination of transmission to humans. J. Med. Microbiol. 63, 399–407 (2014).
Hulin, V. et al. Host preference and zoonotic potential of Chlamydia psittaci and C. gallinacea in poultry. Pathog. Dis. 73, 1–11 (2015).
Kaltenboeck, B., Kousoulas, K. G. & Storz, J. Structures of and allelic diversity and relationships among the major outer membrane protein (ompA) genes of the four chlamydial species. J. Bacteriol. 175, 487–502 (1993).
Mohamad, K. Y. et al. Host adaptation of Chlamydia pecorum towards low virulence evident in co-evolution of the ompA, incA and ORF663 Loci. PLoS One. 9, e103615 (2014).
Reinhold, P., Sachse, K. & Kaltenboeck, B. Chlamydiaceae in cattle: Commensals, trigger organisms, or pathogens? Vet. J. 189, 257–267 (2011).
Poudel, A., Elsasser, T. H., Rahman, K. S., Chowdhury, E. U. & Kaltenboeck, B. Asymptomatic endemic Chlamydia pecorum infections reduce growth rates in calves by up to 48 percent. PLoS One. 7, e44961 (2012).
Wei, L. et al. Use of a universal hydroxymethylbilane synthase (HMBS)-based PCR as an endogenous internal control and to enable typing of mammalian DNAs. Appl. Microbiol. Biotechnol. 98, 5579–5587 (2014).
Kelly, P. J. et al. Ehrlichiosis, babesiosis, anaplasmosis and hepatozoonosis in dogs from St. Kitts, West Indies. PLoS One. 8, e53450 (2013).
Yang, Y. et al. A pan-Theileria FRET-qPCR survey for Theileria spp. in ruminants from nine provinces of China. Parasit. Vectors. 7, 413 (2014).
Laroucau, K. et al. Chlamydial infections in duck farms associated with human cases of psittacosis in France. Vet. Microbiol. 135, 82–89 (2009).
Vorimore, F. et al. Isolation of a new Chlamydia species from the feral Sacred Ibis (Threskiornis aethiopicus): Chlamydia ididis. PLoS One. 8, e74823 (2013).
Acknowledgements
This project was supported by grant from the National Natural Science Foundation of China (NO: 31272575) and the Priority Academic Program Development of Jiangsu Higher Education Institutions, Yangzhou, Jiangsu, P. R. China (www.ec.js.edu.cn/art/2014/3/27/art_11351_147088.html).
Author information
Authors and Affiliations
Contributions
C.W., W.G., J.L. and B.K. conceived and designed the experiments. W.G., J.L., J.G. and W.F. collected samples and performed the experiments. C.W., W.G. and B.K. analyzed the data and C.W., W.G. and B.K. wrote the manuscript. All authors reviewed and approved the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Guo, W., Li, J., Kaltenboeck, B. et al. Chlamydia gallinacea, not C. psittaci, is the endemic chlamydial species in chicken (Gallus gallus). Sci Rep 6, 19638 (2016). https://doi.org/10.1038/srep19638
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep19638
This article is cited by
-
Proof of concept for multiplex detection of antibodies against Chlamydia species in chicken serum using a bead-based suspension array with peptides as antigens
Veterinary Research (2023)
-
Experimental Chlamydia gallinacea infection in chickens does not protect against a subsequent experimental Chlamydia psittaci infection
Veterinary Research (2021)
-
Genetic and phenotypic analysis of the pathogenic potential of two novel Chlamydia gallinacea strains compared to Chlamydia psittaci
Scientific Reports (2021)
-
A comprehensive review on avian chlamydiosis: a neglected zoonotic disease
Tropical Animal Health and Production (2021)
-
Hydroxymethylbilane synthase (HMBS) gene-based endogenous internal control for avian species
AMB Express (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.