Abstract
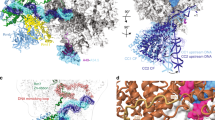
Yeast RNA polymerase II (Pol II) general transcription factor TFIIE and the TFIIH subunit Ssl2 (yeast ortholog of mammalian XPB) function in the transition of the preinitiation complex (PIC) to the open complex. We show that the three TFIIE winged-helix (WH) domains form a heterodimer, with the Tfa1 (TFIIEα) WH binding the Pol II clamp and the Tfa2 (TFIIEβ) tandem WH domain encircling promoter DNA that becomes single-stranded in the open complex. Ssl2 lies adjacent to TFIIE, enclosing downstream promoter DNA. Unlike previous proposals, comparison of the PIC and open-complex models strongly suggests that Ssl2 promotes DNA opening by functioning as a double-stranded-DNA translocase, feeding 15 base pairs into the Pol II cleft. Right-handed threading of DNA through the Ssl2 binding groove, combined with the fixed position of upstream promoter DNA, leads to DNA unwinding and the open state.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Hahn, S. & Young, E.T. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics 189, 705–736 (2011).
Thomas, M.C. & Chiang, C.M. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41, 105–178 (2006).
Wang, W., Carey, M. & Gralla, J.D. Polymerase II promoter activation: closed complex formation and ATP-driven start site opening. Science 255, 450–453 (1992).
Holstege, F.C.P., Fiedler, U. & Timmers, H.T.M. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 16, 7468–7480 (1997).
Chen, H.T., Warfield, L. & Hahn, S. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat. Struct. Mol. Biol. 14, 696–703 (2007).
Eichner, J., Chen, H.T., Warfield, L. & Hahn, S. Position of the general transcription factor TFIIF within the RNA polymerase II transcription preinitiation complex. EMBO J. 29, 706–716 (2010).
Chen, Z.A. et al. Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29, 717–726 (2010).
Fishburn, J. & Hahn, S. Architecture of the yeast RNA polymerase II open complex and regulation of activity by TFIIF. Mol. Cell Biol. 32, 12–25 (2012).
Kostrewa, D. et al. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature 462, 323–330 (2009).
Liu, X., Bushnell, D.A., Wang, D., Calero, G. & Kornberg, R.D. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science 327, 206–209 (2010).
Pan, G. & Greenblatt, J. Initiation of transcription by RNA Polymerase II is limited by melting of the promoter DNA in the region immediately upstream of the initiation site. J. Biol. Chem. 269, 30101–30104 (1994).
Tantin, D. & Carey, M. A heteroduplex template circumvents the energetic requirement for ATP during activated transcription by RNA Pol II. J. Biol. Chem. 269, 17937–17400 (1994).
Holstege, F.C., van der Vliet, P.C. & Timmers, H.T. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 15, 1666–1677 (1996).
Ohkuma, Y. Multiple functions of general transcription factors TFIIE and TFIIH in transcription: possible points of regulation by trans-acting factors. J. Biochem. 122, 481–489 (1997).
Feaver, W.J. et al. Yeast TFIIE. Cloning, expression, and homology to vertebrate proteins. J. Biol. Chem. 269, 27549–27553 (1994).
Ohkuma, Y., Hashimoto, S., Wang, C.K., Horikoshi, M. & Roeder, R.G. Analysis of the role of TFIIE in basal transcription and TFIIH-mediated carboxy-terminal domain phosphorylation through structure-function studies of TFIIE-alpha. Mol. Cell Biol. 15, 4856–4866 (1995).
Okuda, M. et al. A novel zinc finger structure in the large subunit of human general transcription factor TFIIE. J. Biol. Chem. 279, 51395–51403 (2004).
Kuldell, N.H. & Buratowski, S. Genetic analysis of the large subunit of yeast transcription factor IIE reveals two regions with distinct functions. Mol. Cell Biol. 17, 5288–5298 (1997).
Sakurai, H., Ohishi, T. & Fukasawa, T. Promoter structure–dependent functioning of the general transcription factor IIE in Saccharomyces cerevisiae. J. Biol. Chem. 272, 15936–15942 (1997).
Aravind, L., Anantharaman, V., Balaji, S., Babu, M.M. & Iyer, L.M. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 29, 231–262 (2005).
Meinhart, A., Blobel, J. & Cramer, P. An extended winged helix domain in general transcription factor E/IIEα. J. Biol. Chem. 278, 48267–48274 (2003).
Geiger, S.R. et al. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol. Cell 39, 583–594 (2010).
Okuda, M. et al. Structure of the central core domain of TFIIEβ with a novel double-stranded DNA-binding surface. EMBO J. 19, 1346–1356 (2000).
Tanaka, A., Watanabe, T., Iida, Y., Hanaoka, F. & Ohkuma, Y. Central forkhead domain of human TFIIE beta plays a primary role in binding double-stranded DNA at transcription initiation. Genes Cells 14, 395–405 (2009).
Shinkai, A. et al. The putative DNA-binding protein Sto12a from the thermoacidophilic archaeon Sulfolobus tokodaii contains intrachain and interchain disulfide bonds. J. Mol. Biol. 372, 1293–1304 (2007).
Jawhari, A. et al. Structure and oligomeric state of human transcription factor TFIIE. EMBO Rep. 7, 500–505 (2006).
Luo, J., Fishburn, J., Hahn, S. & Ranish, J. An integrated chemical cross-linking and mass spectrometry approach to study protein complex architecture and function. Mol. Cell Proteomics 11, M111.008318 (2012).
Kim, T.K., Ebright, R.H. & Reinberg, D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science 288, 1418–1421 (2000).
Miller, G. & Hahn, S. A DNA-tethered cleavage probe reveals the path for promoter DNA in the yeast preinitiation complex. Nat. Struct. Mol. Biol. 13, 603–610 (2006).
Grohmann, D. et al. The initiation factor TFE and the elongation factor Spt4/5 compete for the RNAP clamp during transcription initiation and elongation. Mol. Cell 43, 263–274 (2011).
Grünberg, S., Bartlett, M.S., Naji, S. & Thomm, M. Transcription factor E is a part of transcription elongation complexes. J. Biol. Chem. 282, 35482–35490 (2007).
Coin, F., Bergmann, E., Tremeau-Bravard, A. & Egly, J.M. Mutations in XPB and XPD helicases found in xeroderma pigmentosum patients impair the transcription function of TFIIH. EMBO J. 18, 1357–1366 (1999).
Lin, Y.C., Choi, W.S. & Gralla, J.D. TFIIH XPB mutants suggest a unified bacterial-like mechanism for promoter opening but not escape. Nat. Struct. Mol. Biol. 12, 603–607 (2005).
Datwyler, S.A. & Meares, C.F. Protein-protein interactions mapped by artificial proteases: where sigma factors bind to RNA polymerase. Trends Biochem. Sci. 25, 408–414 (2000).
Chen, H.T. & Hahn, S. Binding of TFIIB to RNA polymerase II: Mapping the binding site for the TFIIB zinc ribbon domain within the preinitiation complex. Mol. Cell 12, 437–447 (2003).
Ranish, J.A., Yudkovsky, N. & Hahn, S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 13, 49–63 (1999).
Liu, X., Bushnell, D.A., Silva, D.A., Huang, X. & Kornberg, R.D. Initiation complex structure and promoter proofreading. Science 333, 633–637 (2011).
Douziech, M. et al. Mechanism of promoter melting by the xeroderma pigmentosum complementation group B helicase of transcription factor IIH revealed by protein-DNA photo-cross-linking. Mol. Cell Biol. 20, 8168–8177 (2000).
Caruthers, J.M. & McKay, D.B. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12, 123–133 (2002).
Goel, S., Krishnamurthy, S. & Hampsey, M. Mechanism of start site selection by RNA polymerase II: interplay between TFIIB and Ssl2/XPB helicase subunit of TFIIH. J. Biol. Chem. 287, 557–567 (2012).
Chin, J.W. et al. An expanded eukaryotic genetic code. Science 301, 964–967 (2003).
Kettenberger, H., Armache, K.J. & Cramer, P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell 16, 955–965 (2004).
Naji, S., Grunberg, S. & Thomm, M. The RPB7 orthologue E' is required for transcriptional activity of a reconstituted archaeal core enzyme at low temperatures and stimulates open complex formation. J. Biol. Chem. 282, 11047–11057 (2007).
Werner, F. & Grohmann, D. Evolution of multisubunit RNA polymerases in the three domains of life. Nat. Rev. Microbiol. 9, 85–98 (2011).
Vannini, A. & Cramer, P. Conservation between the RNA Polymerase I, II, and III Transcription Initiation Machineries. Mol. Cell 45, 439–446 (2012).
Bischler, N. et al. Localization of the yeast RNA polymerase I–specific subunits. EMBO J. 21, 4136–4144 (2002).
Wu, C.C., Lin, Y.C. & Chen, H.T. The TFIIF-like Rpc37/53 dimer lies at the center of a protein network to connect TFIIIC, Bdp1, and the RNA polymerase III active center. Mol. Cell Biol. 31, 2715–2728 (2011).
Jennebach, S., Herzog, F., Aebersold, R. & Cramer, P. Crosslinking-MS analysis reveals RNA polymerase I domain architecture and basis of rRNA cleavage. Nucleic Acids Res. published online, doi:10.1093/nar/gks220 (6 March 2012).
Okamoto, T. et al. Analysis of the role of TFIIE in transcriptional regulation through structure-function studies of the TFIIEβ subunit. J. Biol. Chem. 273, 19866–19876 (1998).
Brun, I., Sentenac, A. & Werner, M. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J. 16, 5730–5741 (1997).
Vannini, A. et al. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell 143, 59–70 (2010).
Coin, F. et al. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat. Genet. 20, 184–188 (1998).
Lin, Y.C. & Gralla, J.D. Stimulation of the XPB ATP-dependent helicase by the beta subunit of TFIIE. Nucleic Acids Res. 33, 3072–3081 (2005).
Treutlein, B. et al. Dynamic architecture of a minimal RNA polymerase II open promoter complex. Mol. Cell 46, 136–146 (2012).
Söding, J., Biegert, A. & Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 (2005).
Eswar, N., Eramian, D., Webb, B., Shen, M.Y. & Sali, A. Protein structure modeling with MODELLER. Methods Mol. Biol. 426, 145–159 (2008).
Colovos, C. & Yeates, T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 2, 1511–1519 (1993).
Lüthy, R., Bowie, J.U. & Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature 356, 83–85 (1992).
Davis, I.W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Acknowledgements
We thank J. Luo and J. Ranish (Institute for Systems Biology, Seattle, Washington) for sharing their unpublished TFIIE–cross-linking data, J. Fishburn for advice and help with TFIIE purification and transcription assays, A. Vannini for coordinates of the Rpc34 WH model, G. Smith for discussions and B. Knutson, I. Kamenova and M. Bartlett for comments on the manuscript. This work was supported by grant RO1GM053451 to S.H. and Forschungsstipendium GR 3776/2-1 of the Deutsche Forschungsgemeinschaft to S.G.
Author information
Authors and Affiliations
Contributions
S.G. designed and performed all biochemical experiments except for the Bpa cross-linking, which was designed and performed by L.W.; S.G. and S.H. designed and performed Tfa1 and Tfa2 genetic assays, performed structure modeling and prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–5 and Supplementary Tables 1 and 2 (PDF 3323 kb)
Supplementary Movie 1
Overall structure of the PIC and open complex models. (MOV 6594 kb)
Rights and permissions
About this article
Cite this article
Grünberg, S., Warfield, L. & Hahn, S. Architecture of the RNA polymerase II preinitiation complex and mechanism of ATP-dependent promoter opening. Nat Struct Mol Biol 19, 788–796 (2012). https://doi.org/10.1038/nsmb.2334
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2334
This article is cited by
-
Structures of mammalian RNA polymerase II pre-initiation complexes
Nature (2021)
-
Organization and regulation of gene transcription
Nature (2019)
-
Transcription preinitiation complex structure and dynamics provide insight into genetic diseases
Nature Structural & Molecular Biology (2019)
-
TFIIE orchestrates the recruitment of the TFIIH kinase module at promoter before release during transcription
Nature Communications (2019)
-
Structures of transcription pre-initiation complex with TFIIH and Mediator
Nature (2017)