Abstract
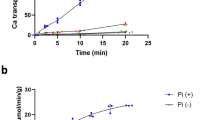
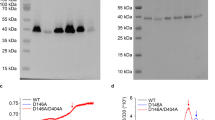
Active exchangers dissipate the gradient of one substrate to accumulate nutrients, export xenobiotics and maintain cellular homeostasis. Mechanistic studies have suggested that two fundamental properties are shared by all exchangers: substrate binding is antagonistic, and coupling is maintained by preventing shuttling of the empty transporter. The CLC H+/Cl− exchangers control the homeostasis of cellular compartments in most living organisms, but their transport mechanism remains unclear. We show that substrate binding to CLC-ec1 is synergistic rather than antagonistic: chloride binding induces protonation of a crucial glutamate. The simultaneous binding of H+ and Cl− gives rise to a fully loaded state that is incompatible with conventional transport mechanisms. Mutations in the Cl− transport pathway identically alter the stoichiometries of H+/Cl− exchange and binding. We propose that the thermodynamics of synergistic substrate binding, rather than the kinetics of conformational changes and ion binding, determine the stoichiometry of transport.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Jardetzky, O. Simple allosteric model for membrane pumps. Nature 211, 969–970 (1966).
Jennings, M.L. Structure and function of the red blood cell anion transport protein. Annu. Rev. Biophys. Biophys. Chem. 18, 397–430 (1989).
Arkin, I.T. et al. Mechanism of Na+/H+ antiporting. Science 317, 799–803 (2007).
Khare, D., Oldham, M.L., Orelle, C., Davidson, A.L. & Chen, J. Alternating access in maltose transporter mediated by rigid-body rotations. Mol. Cell 33, 528–536 (2009).
Shimamura, T. et al. Molecular basis of alternating access membrane transport by the sodium-hydantoin transporter Mhp1. Science 328, 470–473 (2010).
Kaback, H.R., Smirnova, I., Kasho, V., Nie, Y. & Zhou, Y. The alternating access transport mechanism in LacY. J. Membr. Biol. 239, 85–93 (2011).
Radestock, S. & Forrest, L.R. The alternating-access mechanism of MFS transporters arises from inverted-topology repeats. J. Mol. Biol. 407, 698–715 (2011).
Milanick, M.A. Ferret red cells: Na/Ca exchange and Na-K-Cl cotransport. Comp. Biochem. Physiol. Comp. Physiol. 102, 619–624 (1992).
Sekler, I., Lo, R.S. & Kopito, R.R. A conserved glutamate is responsible for ion selectivity and pH dependence of the mammalian anion exchangers AE1 and AE2. J. Biol. Chem. 270, 28751–28758 (1995).
Beaugé, L. & DiPolo, R. The squid axon Na+/Ca2+ exchanger shows ping-pong kinetics only when the Cai-regulatory site is saturated. Cell. Physiol. Biochem. 23, 37–42 (2009).
Adam, Y., Tayer, N., Rotem, D., Schreiber, G. & Schuldiner, S. The fast release of sticky protons: kinetics of substrate binding and proton release in a multidrug transporter. Proc. Natl. Acad. Sci. USA 104, 17989–17994 (2007).
Restrepo, D., Cronise, B.L., Snyder, R.B. & Knauf, P.A. A novel method to differentiate between ping-pong and simultaneous exchange kinetics and its application to the anion exchanger of the HL60 cell. J. Gen. Physiol. 100, 825–846 (1992).
Contreras-Jurado, C., Sanchez-Morito, N., Ruiz-Contreras, A., Gonzalez-Martinez, M.T. & Soler-Diaz, A. Evidence for simultaneous 1Na+:1Mg2+ and ping-pong 2Na+:1Mg2+ exchangers in rat thymocyte. Front. Biosci. 10, 1693–1706 (2005).
Dutzler, R., Campbell, E.B., Cadene, M., Chait, B.T. & MacKinnon, R. X-ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature 415, 287–294 (2002).
Abramson, J. et al. Structure and mechanism of the lactose permease of Escherichia coli. Science 301, 610–615 (2003).
Yernool, D., Boudker, O., Jin, Y. & Gouaux, E. Structure of a glutamate transporter homolog from Pyrococcus horikoshii. Nature 431, 811–818 (2004).
Yamashita, A., Singh, S.K., Kawate, T., Jin, Y. & Gouaux, E. Crystal structure of a bacterial homolog of Na+/Cl−-dependent neurotransmitter transporters. Nature 437, 215–223 (2005).
Morth, J.P. et al. Crystal structure of the sodium-potassium pump. Nature 450, 1043–1049 (2007).
Olesen, C. et al. The structural basis of calcium transport by the calcium pump. Nature 450, 1036–1042 (2007).
Fang, Y. et al. Structure of a prokaryotic virtual proton pump at 3.2-Å resolution. Nature 460, 1040–1043 (2009).
Shaffer, P.L., Goehring, A., Shankaranarayanan, A. & Gouaux, E. Structure and mechanism of a Na+-independent amino acid transporter. Science 325, 1010–1014 (2009).
Accardi, A. & Miller, C. Secondary active transport mediated by a prokaryotic homolog of ClC Cl− channels. Nature 427, 803–807 (2004).
Scheel, O., Zdebik, A.A., Lourdel, S. & Jentsch, T.J. Voltage-dependent electrogenic chloride/proton exchange by endosomal CLC proteins. Nature 436, 424–427 (2005).
Picollo, A. & Pusch, M. Chloride/proton antiporter activity of mammalian CLC proteins ClC-4 and ClC-5. Nature 436, 420–423 (2005).
De Angeli, A. et al. The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature 442, 939–942 (2006).
Graves, A.R., Curran, P., Smith, C. & Mindell, J. The Cl−/H+ antiporter ClC-7 is the primary chloride permeation pathway in lysosomes. Nature 453, 788 (2008).
Matsuda, J.J. et al. Overexpression of CLC-3 in HEK293T cells yields novel currents that are pH dependent. Am. J. Physiol. Cell Physiol. 294, C251–262 (2008).
Accardi, A. et al. Separate ion pathways in a Cl−/H+ exchanger. J. Gen. Physiol. 126, 563–570 (2005).
Feng, L., Campbell, E.B., Hsiung, Y. & MacKinnon, R. Structure of a eukaryotic CLC transporter defines an intermediate state in the transport cycle. Science 330, 635–641 (2010).
Zdebik, A.A. et al. Determinants of anion-proton coupling in mammalian endosomal CLC proteins. J. Biol. Chem. 283, 4219–4227 (2008).
Dutzler, R., Campbell, E.B. & MacKinnon, R. Gating the selectivity filter in ClC chloride channels. Science 300, 108–112 (2003).
Accardi, A. & Picollo, A. CLC channels and transporters: proteins with borderline personalities. Biochim. Biophys. Acta 1798, 1457–1464 (2010).
Novarino, G., Weinert, S., Rickheit, G. & Jentsch, T.J. Endosomal chloride-proton exchange rather than chloride conductance is crucial for renal endocytosis. Science 328, 1398–1401 (2010).
Baker, B.M. & Murphy, K.P. Evaluation of linked protonation effects in protein binding reactions using isothermal titration calorimetry. Biophys. J. 71, 2049–2055 (1996).
Petrosian, S.A. & Makhatadze, G.I. Contribution of proton linkage to the thermodynamic stability of the major cold-shock protein of Escherichia coli CspA. Protein Sci. 9, 387–394 (2000).
Velazquez-Campoy, A. et al. Thermodynamic dissection of the binding energetics of KNI-272, a potent HIV-1 protease inhibitor. Protein Sci. 9, 1801–1809 (2000).
Picollo, A., Malvezzi, M., Houtman, J. & Accardi, A. Basis of substrate binding and conservation of selectivity in the CLC family of channels and transporters. Nat. Struct. Mol. Biol. 16, 1294–1301 (2009).
Accardi, A., Lobet, S., Williams, C., Miller, C. & Dutzler, R. Synergism between halide binding and proton transport in a CLC-type exchanger. J. Mol. Biol. 362, 691–699 (2006).
Walden, M. et al. Uncoupling and turnover in a Cl−/H+ exchange transporter. J. Gen. Physiol. 129, 317–329 (2007).
Iyer, R., Iverson, T.M., Accardi, A. & Miller, C. A biological role for prokaryotic ClC chloride channels. Nature 419, 715–718 (2002).
Accardi, A., Kolmakova-Partensky, L., Williams, C. & Miller, C. Ionic currents mediated by a prokaryotic homolog of CLC Cl− channels. J. Gen. Physiol. 123, 109–119 (2004).
Nguitragool, W. & Miller, C. Uncoupling of a CLC Cl−/H+ exchange transporter by polyatomic anions. J. Mol. Biol. 362, 682–690 (2006).
Lobet, S. & Dutzler, R. Ion-binding properties of the ClC chloride selectivity filter. EMBO J. 25, 24–33 (2006).
Jenks, W.P. Catalysis in Chemistry and Enzymology. (McGraw Hill Text, 1969).
Zifarelli, G. & Pusch, M. Conversion of the 2 Cl−/1 H+ antiporter ClC-5 in a NO3−/H+ antiporter by a single point mutation. EMBO J. 28, 175–182 (2009).
Richard, E.A. & Miller, C. Steady-state coupling of ion-channel conformations to a transmembrane ion gradient. Science 247, 1208–1210 (1990).
Lísal, J. & Maduke, M. The ClC-0 chloride channel is a 'broken' Cl−/H+ antiporter. Nat. Struct. Mol. Biol. 15, 805–810 (2008).
Chen, M.F. & Chen, T.Y. Side-chain charge effects and conductance determinants in the pore of ClC-0 chloride channels. J. Gen. Physiol. 122, 133–145 (2003).
Matsuda, J.J., Filali, M., Collins, M., Volk, K. & Lamb, F. The ClC-3 Cl−/H+ antiporter becomes uncoupled at low extracellular pH. J. Biol. Chem. 285, 2569–2579 (2010).
Thomson, J.A. & Ladbury, J.E. in Biocalorimetry 2: Applications of Calorimetry in the Biological Sciences (eds. Ladbury, J.E. & Doyle, M.L.) Ch. 2 (Wiley, 2004).
Fukada, H. & Takahashi, K. Enthalpy and heat capacity changes for the proton dissociation of various buffer components in 0.1 M potassium chloride. Proteins 33, 159–166 (1998).
Maduke, M., Pheasant, D.J. & Miller, C. High-level expression, functional reconstitution, and quaternary structure of a prokaryotic ClC-type chloride channel. J. Gen. Physiol. 114, 713–722 (1999).
Jo, S., Kim, T., Iyer, V.G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Brooks, B.R. et al. CHARMM: the biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 (2009).
MacKerell, A.D. Jr. et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998).
Boiteux, C. & Bernèche, S. Absence of ion-binding affinity in the putatively inactivated low-[K+] structure of the KcsA potassium channel. Structure 19, 70–79 (2011).
Souaille, M. & Roux, B. Extension to the weighted histogram analysis method: combining umbrella sampling with free energy calculations. Comput. Phys. Commun. 135, 40–57 (2001).
Shirts, M.R. & Chodera, J.D. Statistically optimal analysis of samples from multiple equilibrium states. J. Chem. Phys. 129, 124105 (2008).
Pohorille, A., Jarzynski, C. & Chipot, C. Good practices in free-energy calculations. J. Phys. Chem. B 114, 10235–10253 (2010).
Faraldo-Gómez, J.D. & Roux, B. Electrostatics of ion stabilization in a ClC chloride channel homolog from Escherichia coli. J. Mol. Biol. 339, 981–1000 (2004).
Acknowledgements
The authors thank C. Miller (Brandeis University) for the gift of 36Cl− and D. Posson, D. Basilio, M. Malvezzi, C. Nimigean and H. Weinstein for helpful discussions and comments on the manuscript. This work was supported by grants from the National Institutes of Health (1R01GM085232 to A.A.) and by a grant from the Swiss National Science Foundation (SNF-Professorship #118928 to S.B.). Y.X. is supported by the China Scholarship Council. Part of the simulations were performed using the facilities of the Swiss National Supercomputing Centre (CSCS).
Author information
Authors and Affiliations
Contributions
A.P. performed the ITC measurements; A.A. and A.P. performed the flux experiments and analyzed data; Y.X., N.J. and S.B. performed and analyzed the simulations; A.A. designed research and wrote the paper; all authors contributed to the editing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Tables
Supplementary Figures 1–4 and Supplementary Tables 1–2 (PDF 2230 kb)
Rights and permissions
About this article
Cite this article
Picollo, A., Xu, Y., Johner, N. et al. Synergistic substrate binding determines the stoichiometry of transport of a prokaryotic H+/Cl− exchanger. Nat Struct Mol Biol 19, 525–531 (2012). https://doi.org/10.1038/nsmb.2277
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.2277
This article is cited by
-
Ion and lipid orchestration of secondary active transport
Nature (2024)
-
A CLC-type F-/H+ antiporter in ion-swapped conformations
Nature Structural & Molecular Biology (2018)
-
The Ec-NhaA antiporter switches from antagonistic to synergistic antiport upon a single point mutation
Scientific Reports (2016)
-
Conformational changes required for H+/Cl− exchange mediated by a CLC transporter
Nature Structural & Molecular Biology (2014)
-
Fluoride-dependent interruption of the transport cycle of a CLC Cl−/H+ antiporter
Nature Chemical Biology (2013)