Abstract
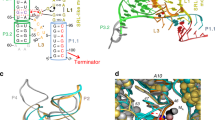
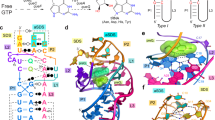
The second messenger signaling molecule bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP) regulates many processes in bacteria, including motility, pathogenesis and biofilm formation. c-di-GMP–binding riboswitches are important downstream targets in this signaling pathway. Here we report the crystal structure, at 2.7 Å resolution, of a c-di-GMP riboswitch aptamer from Vibrio cholerae bound to c-di-GMP, showing that the ligand binds within a three-helix junction that involves base-pairing and extensive base-stacking. The symmetric c-di-GMP is recognized asymmetrically with respect to both the bases and the backbone. A mutant aptamer was engineered that preferentially binds the candidate signaling molecule c-di-AMP over c-di-GMP. Kinetic and structural data suggest that genetic regulation by the c-di-GMP riboswitch is kinetically controlled and that gene expression is modulated through the stabilization of a previously unidentified P1 helix, illustrating a direct mechanism for c-di-GMP signaling.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Hengge, R. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7, 263–273 (2009).
Römling, U. & Amikam, D. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 9, 218–228 (2006).
Tamayo, R., Pratt, J.T. & Camilli, A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 61, 131–148 (2007).
Waters, C.M., Lu, W., Rabinowitz, J.D. & Bassler, B. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 190, 2527–2536 (2008).
Fong, J.C. & Yildiz, F. Interplay between cyclic AMP-cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholerae biofilm formation. J. Bacteriol. 190, 6646–6659 (2008).
Jenal, U. & Malone, J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40, 385–407 (2006).
Ryan, R., Fouhy, Y., Lucey, J. & Dow, J.M. Cyclic di-GMP signaling in bacteria: recent advances and new puzzles. J. Bacteriol. 188, 8327–8334 (2006).
Sudarsan, N. et al. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321, 411–413 (2008).
Roth, A. & Breaker, R.R. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 78, 305–334 (2009).
Lim, B., Beyhan, S., Meir, J. & Yildiz, F. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60, 331–348 (2006).
Beyhan, S. & Yildiz, F. Smooth to rugose phase variation in Vibrio cholerae can be mediated by a single nucleotide change that targets c-di-GMP signalling pathway. Mol. Microbiol. 63, 995–1007 (2007).
Ferré-D'Amaré, A.R. & Doudna, J.A. Crystallization and structure determination of a hepatitis delta virus ribozyme: use of the RNA-binding protein U1A as a crystallization module. J. Mol. Biol. 295, 541–556 (2000).
Galsbøl, F.H. & Simonsen, K. The preparation, separation and characterization of some ammine complexes of Iridium(III). Acta Chem. Scand. 44, 796–801 (1990).
Wickiser, J.K., Cheah, M., Breaker, R. & Crothers, D.M. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry 44, 13404–13414 (2005).
Wickiser, J.K., Winkler, W., Breaker, R. & Crothers, D.M. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell 18, 49–60 (2005).
Witte, G., Hartung, S., Büttner, K. & Hopfner, K.P. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol. Cell 30, 167–178 (2008).
Römling, U. Great times for small molecules: c-di-AMP, a second messenger candidate in Bacteria and Archaea. Sci. Signal. 1, pe39 (2008).
Amikam, D. & Galperin, M. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22, 3–6 (2006).
Ryjenkov, D.A., Simm, R., Römling, U. & Gomelsky, M. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 281, 30310–30314 (2006).
Benach, J. et al. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 26, 5153–5166 (2007).
Christen, M. et al. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 104, 4112–4117 (2007).
Merighi, M., Lee, V., Hyodo, M., Hayakawa, Y. & Lory, S. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65, 876–895 (2007).
Pratt, J.T., Tamayo, R., Tischler, A. & Camilli, A. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 282, 12860–12870 (2007).
Minasov, G. et al. Crystal structures of YkuI and its complex with second messenger c-di-GMP suggests catalytic mechanism of phosphodiester bond cleavage by EAL domains. J. Biol. Chem. 284, 13174–13184 (2009).
Barends, T.R.M. et al. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 459, 1015–1018 (2009).
Chan, C. et al. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. USA 101, 17084–17089 (2004).
Wassmann, P. et al. Structure of BeF3- -modified response regulator PleD: implications for diguanylate cyclase activation, catalysis, and feedback inhibition. Structure 15, 915–927 (2007).
De, N. et al. Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol. 6, e67 (2008).
Winkler, W., Cohen-Chalamish, S. & Breaker, R. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. USA 99, 15908–15913 (2002).
Mandal, M., Boese, B., Barrick, J., Winkler, W. & Breaker, R. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113, 577–586 (2003).
Winkler, W., Nahvi, A., Sudarsan, N., Barrick, J. & Breaker, R. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 10, 701–707 (2003).
Sudarsan, N. et al. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes. Dev. 17, 2688–2697 (2003).
Leontis, N.B. & Westhof, E. Geometric nomenclature and classification of RNA base pairs. RNA 7, 499–512 (2001).
Cochrane, J.C., Lipchock, S.V. & Strobel, S. Structural investigation of the GlmS ribozyme bound to its catalytic cofactor. Chem. Biol. 14, 97–105 (2007).
Hyodo, M. & Hayakawa, Y. An improved method for synthesizing cyclic bis(3′–5′)diguanylic acid (c-di-GMP). Bull. Chem. Soc. Jpn. 77, 2089–2093 (2004).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
McCoy, A.J., Grosse-Kunstleve, R.W., Storoni, L.C. & Read, R.J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 (2005).
Terwilliger, T.C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 (1999).
Terwilliger, T.C. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 (2000).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Winn, M.D., Isupov, M.N. & Murshudov, G.N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 (2001).
Brunger, A.T. et al. Crystallography & NMR System (CNS), a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Brunger, A.T. Version 1.2 of the Crystallography and NMR System. Nat. Protoc. 2, 2728–2733 (2007).
DeLano, W.L. The Pymol Molecular Graphics System. http://pymol.org (2008).
Paul, R. et al. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Gend. Dev. 18, 715–727 (2004).
Christen, M., Christen, B., Folcher, M., Schauerte, A. & Jenal, U. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 280, 30829–30837 (2005).
Acknowledgements
We thank S. Myers and the beamline staff at X25, X12C and X29 at the National Synchrotron Light Source at Brookhaven National Laboratory; M. Strickler, D. Keller, and the Yale Center for Structural Biology core staff; Y. Xiong for data processing help and advice; C. Shanahan, E. Butler, N. Carrasco, J. Cochrane and M. Stahley for help and advice; D. Hiller, J. Davis, E. Lee, N. Sudarsan and other members of the Strobel and Breaker labs for helpful discussions; V. Singh (Strobel lab) for synthesis of iridium hexammine; U. Jenal (University of Basel) for the gift of the PleD* expression plasmid; K.-P. Hopfner (University of Munich) for the gift of the DisA expression plasmid; and G. Reguera (Michigan State University) and B. Bassler (Princeton University) for the gift of V. cholerae genomic DNA. This work was supported by the US National Institutes of Health grant GM02278 and the National Science Foundation grant MCB0544255 to S.A.S.
Author information
Authors and Affiliations
Contributions
K.D.S. determined the crystal structure, performed biochemical experiments and wrote the manuscript. S.V.L. performed biochemical experiments. T.D.A. performed the updated sequence alignment and performed the in-line probing experiments. J.W. gave critical help in data processing and refinement. R.R.B. and S.A.S. advised on the project. All authors discussed the data and provided comments on the manuscript.
Corresponding author
Ethics declarations
Competing interests
R.R.B. is a cofounder of BioRelix, a biotechnology company pursuing the development of riboswitches as antibiotics targets.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7, Supplementary Data/Discussion and Supplementary Methods (PDF 4959 kb)
Rights and permissions
About this article
Cite this article
Smith, K., Lipchock, S., Ames, T. et al. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat Struct Mol Biol 16, 1218–1223 (2009). https://doi.org/10.1038/nsmb.1702
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1702
This article is cited by
-
Structure and mechanism of a methyltransferase ribozyme
Nature Chemical Biology (2022)
-
The fluorescent aptamer Squash extensively repurposes the adenine riboswitch fold
Nature Chemical Biology (2022)
-
Local-to-global signal transduction at the core of a Mn2+ sensing riboswitch
Nature Communications (2019)
-
BrlR from Pseudomonas aeruginosa is a receptor for both cyclic di-GMP and pyocyanin
Nature Communications (2018)
-
Computational and NMR spectroscopy insights into the conformation of cyclic di-nucleotides
Scientific Reports (2017)