Key Points
-
Spectroscopically directed metabolic profiling has the potential to enhance current capabilities in patient stratification
-
Examples of the contribution of metabolic profiling to disease diagnostics and prognostics as applied to liver and digestive diseases are widely present in the literature
-
Metabolic profiling provides insights about the function of gut bacteria, which are known to be involved in a wide range of pathologies including IBD, obesity, fatty liver and colorectal cancer
-
Surgical metabonomics centred on mass-spectrometry-linked intelligent surgical devices have the potential to deliver clinically relevant chemical information in real-time to augment clinical decision-making
Abstract
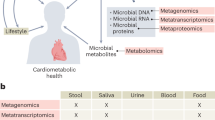
Disease risk and treatment response are determined, at the individual level, by a complex history of genetic and environmental interactions, including those with our endogenous microbiomes. Personalized health care requires a deep understanding of patient biology that can now be measured using a range of '-omics' technologies. Patient stratification involves the identification of genetic and/or phenotypic disease subclasses that require different therapeutic strategies. Stratified medicine approaches to disease diagnosis, prognosis and therapeutic response monitoring herald a new dimension in patient care. Here, we explore the potential value of metabolic profiling as applied to unmet clinical needs in gastroenterology and hepatology. We describe potential applications in a number of diseases, with emphasis on large-scale population studies as well as metabolic profiling on the individual level, using spectrometric and imaging technologies that will leverage the discovery of mechanistic information and deliver novel health care solutions to improve clinical pathway management.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Nicholson, J. K. Global systems biology, personalized medicine and molecular epidemiology. Mol. Syst. Biol. 2, 52 (2006).
Ogino, S. et al. Interdisciplinary education to integrate pathology and epidemiology: towards molecular and population-level health science. Am. J. Epidemiol. 176, 659–667 (2012).
Kalow, W. Pharmacogenetics and personalised medicine. Fundam. Clin. Pharmacol. 16, 337–342 (2002).
Nicholson, J. K. et al. Metabolic phenotyping in clinical and surgical environments. Nature 491, 384–392 (2012).
Holmes, E., Wilson, I. D. & Nicholson, J. K. Metabolic phenotyping in health and disease. Cell 134, 714–717 (2008).
Hood, L., Balling, R. & Auffray, C. Revolutionizing medicine in the 21st century through systems approaches. Biotechnol. J. 7, 992–1001 (2012).
Pimiento, J. M. et al. Melanoma genotypes and phenotypes get personal. Lab. Invest. 93, 858–867 (2013).
Nicholson, J. K., Holmes, E. & Wilson, I. D. Gut microorganisms, mammalian metabolism and personalized health care. Nat. Rev. Microbiol. 3, 431–438 (2005).
Salazar, N. et al. Inulin-type fructans modulate intestinal Bifidobacterium species populations and decrease fecal short-chain fatty acids in obese women. Clin. Nutr. 34, 501-507 (2014).
Faith, J. J., Ahern, P. P., Ridaura, V. K., Cheng, J. & Gordon, J. I. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci. Transl. Med. 6, 220ra11 (2014).
Borgan, E. et al. Subtype-specific response to bevacizumab is reflected in the metabolome and transcriptome of breast cancer xenografts. Mol. Oncol. 7, 130–142 (2013).
Robinette, S. L., Lindon, J. C. & Nicholson, J. K. Statistical spectroscopic tools for biomarker discovery and systems medicine. Anal. Chem. 85, 5297–5303 (2013).
Ciccarelli, O. et al. Pathogenesis of multiple sclerosis: insights from molecular and metabolic imaging. Lancet Neurol. 13, 807–822 (2014).
Woo, J., Baumann, A. & Arguello, V. Recent advancements of flow cytometry: new applications in hematology and oncology. Expert Rev. Mol. Diagn. 14, 67–81 (2014).
Wang, Y. W. et al. Rapid ratiometric biomarker detection with topically applied SERS nanoparticles. Technology (Singap. World Sci.) 2, 118–132 (2014).
Holmes, E. et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 453, 396–400 (2008).
Whiley, L. et al. Evidence of altered phosphatidylcholine metabolism in Alzheimer's disease. Neurobiol. Aging 35, 271–278 (2014).
Maher, A. D. et al. Optimization of human plasma 1H NMR spectroscopic data processing for high-throughput metabolic phenotyping studies and detection of insulin resistance related to type 2 diabetes. Anal. Chem. 80, 7354–7362 (2008).
Hazen, S. L. New lipid and lipoprotein targets for the treatment of cardiometabolic diseases. J. Lipid. Res. 53, 1719–1721 (2012).
Cooper-Dehoff, R. M. et al. Is diabetes mellitus-linked amino acid signature associated with β-blocker-induced impaired fasting glucose? Circ. Cardiovasc. Genet. 7, 199–205 (2014).
Shariff, M. I. et al. Urinary metabolic biomarkers of hepatocellular carcinoma in an Egyptian population: a validation study. J. Proteome Res. 10, 1828–1836 (2011).
Nicholson, J. K., Everett, J. R. & Lindon, J. C. Longitudinal pharmacometabonomics for predicting patient responses to therapy: drug metabolism, toxicity and efficacy. Expert Opin. Drug Metab. Toxicol. 8, 135–139 (2012).
Klootwijk, E. D. et al. Mistargeting of peroxisomal EHHADH and inherited renal Fanconi's syndrome. N. Engl. J. Med. 370, 129–138 (2014).
Pan, Z. et al. Principal component analysis of urine metabolites detected by NMR and DESI-MS in patients with inborn errors of metabolism. Anal. Bioanal. Chem. 387, 539–549 (2007).
Plumb, R. et al. Ultra-performance liquid chromatography coupled to quadrupole-orthogonal time-of-flight mass spectrometry. Rapid Commun. Mass. Spectrom. 18, 2331–2337 (2004).
Shockcor, J. P. et al. Combined HPLC, NMR spectroscopy, and ion-trap mass spectrometry with application to the detection and characterization of xenobiotic and endogenous metabolites in human urine. Anal. Chem. 68, 4431–4435 (1996).
Duarte, I. F., Lamego, I., Rocha, C. & Gil, A. M. NMR metabonomics for mammalian cell metabolism studies. Bioanalysis 1, 1597–1614 (2009).
Jimenez, B. et al. 1H HR-MAS NMR spectroscopy of tumor-induced local metabolic “field-effects” enables colorectal cancer staging and prognostication. J. Proteome Res. 12, 959–968 (2013).
Rocha, C. M. et al. NMR metabolomics of human lung tumours reveals distinct metabolic signatures for adenocarcinoma and squamous cell carcinoma. Carcinogenesis 36, 68–75 (2014).
Veselkov, K. A. et al. Chemo-informatic strategy for imaging mass spectrometry-based hyperspectral profiling of lipid signatures in colorectal cancer. Proc. Natl Acad. Sci. USA 111, 1216–1221 (2014).
Takats, Z., Wiseman, J. M., Gologan, B. & Cooks, R. G. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science 306, 471–473 (2004).
Crecelius, A. C. et al. Three-dimensional visualization of protein expression in mouse brain structures using imaging mass spectrometry. J. Am. Soc. Mass Spectrom. 16, 1093–1099 (2005).
Balluff, B. et al. De novo discovery of phenotypic intratumour heterogeneity using imaging mass spectrometry. J. Pathol. 235, 3–13 (2014).
Veselkov, K. A. et al. Recursive segment-wise peak alignment of biological 1H NMR spectra for improved metabolic biomarker recovery. Anal. Chem. 81, 56–66 (2009).
Vu, T. N. & Laukens, K. Getting your peaks in line: a review of alignment methods for NMR spectral data. Metabolites 3, 259–276 (2013).
Crutchfield, C. A. et al. Comprehensive analysis of LC/MS data using pseudocolor plots. J. Am. Soc. Mass Spectrom. 24, 230–237 (2013).
Trygg, J., Holmes, E. & Lundstedt, T. Chemometrics in metabonomics. J. Proteome Res. 6, 469–479 (2007).
Madsen, R., Lundstedt, T. & Trygg, J. Chemometrics in metabolomics--a review in human disease diagnosis. Anal. Chim. Acta. 659, 23–33 (2010).
Doeswijk, T. G., Smilde, A. K., Hageman, J. A., Westerhuis, J. A. & van Eeuwijk, F. A. On the increase of predictive performance with high-level data fusion. Anal. Chim. Acta. 705, 41–47 (2011).
Cloarec, O. et al. Evaluation of the orthogonal projection on latent structure model limitations caused by chemical shift variability and improved visualization of biomarker changes in 1H NMR spectroscopic metabonomic studies. Anal. Chem. 77, 517–526 (2005).
Posma, J. M., Robinette, S. L., Holmes, E. & Nicholson, J. K. MetaboNetworks, an interactive Matlab-based toolbox for creating, customizing and exploring sub-networks from KEGG. Bioinformatics 30, 893–895 (2014).
Barupal, D. K. et al. MetaMapp: mapping and visualizing metabolomic data by integrating information from biochemical pathways and chemical and mass spectral similarity. BMC Bioinformatics 13, 99 (2012).
Trygg, J. & Wold, S. O2-PLS, a two-block (X-Y) latent variable regression (LVR) method with an integral OSC filter. J. Chemom. 17, 53–64 (2003).
Jayaseelan, K. V. & Steinbeck, C. Building blocks for automated elucidation of metabolites: natural product-likeness for candidate ranking. BMC bioinformatics 15, 234 (2014).
Acar, E., Lawaetz, A. J., Rasmussen, M. A. & Bro, R. Structure-revealing data fusion model with applications in metabolomics. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2013, 6023–6026 (2013).
Bro, R. et al. Data fusion in metabolomic cancer diagnostics. Metabolomics 9, 3–8 (2013).
Vaughan, A. A. et al. Liquid chromatography-mass spectrometry calibration transfer and metabolomics data fusion. Anal. Chem. 84, 9848–9857 (2012).
Cloarec, O. et al. Statistical total correlation spectroscopy: an exploratory approach for latent biomarker identification from metabolic 1H NMR data sets. Anal. Chem. 77, 1282–1289 (2005).
Keun, H. C. et al. Heteronuclear 19F-1H statistical total correlation spectroscopy as a tool in drug metabolism: study of flucloxacillin biotransformation. Anal. Chem. 80, 1073–1079 (2008).
Posma, J. M. et al. Subset optimization by reference matching (STORM): an optimized statistical approach for recovery of metabolic biomarker structural information from 1H NMR spectra of biofluids. Anal. Chem. 84, 10694–10701 (2012).
Shariff, M. I. et al. Hepatocellular carcinoma: current trends in worldwide epidemiology, risk factors, diagnosis and therapeutics. Expert Rev. Gastroenterol. Hepatol. 3, 353–367 (2009).
Wallace, M. C., Preen, D., Jeffrey, G. P. & Adams, L. A. The evolving epidemiology of hepatocellular carcinoma: a global perspective. Expert Rev. Gastroenterol. Hepatol. 9, 765–779 (2015).
Duricova, D., Burisch, J., Jess, T., Gower-Rousseau, C. & Lakatos, P. L. Age-related differences in presentation and course of inflammatory bowel disease: an update on the population-based literature. J. Crohns Colitis 8, 1351–1361 (2014).
O'Keefe, S. J. et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat. Commun. 6, 6342 (2015).
Cho, I. & Blaser, M. J. The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270 (2012).
Dominguez-Bello, M. G., Blaser, M. J., Ley, R. E. & Knight, R. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology 140, 1713–1719 (2011).
Joyce, S. A. & Gahan, C. G. The gut microbiota and the metabolic health of the host. Curr. Opin. Gastroenterol. 30, 120–127 (2014).
Nicholson, J. K. et al. Host-gut microbiota metabolic interactions. Science 336, 1262–1267 (2012).
Pande, C., Kumar, A. & Sarin, S. K. Small-intestinal bacterial overgrowth in cirrhosis is related to the severity of liver disease. Aliment. Pharmacol. Ther. 29, 1273–1281 (2009).
Vonlaufen, A., Spahr, L., Apte, M. V. & Frossard, J. L. Alcoholic pancreatitis: a tale of spirits and bacteria. World J. Gastrointest. Pathophysiol. 5, 82–90 (2014).
Mehal, W. Z. The gordian knot of dysbiosis, obesity and NAFLD. Nat. Rev. Gastroenterol. Hepatol. 10, 637–644 (2013).
Swann, J. R. et al. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc. Natl Acad. Sci. USA 108, 4523–4530 (2011).
Claus, S. P. et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol. Syst. Biol. 4, 219 (2008).
Tlaskalova-Hogenova, H. et al. Microbiome and colorectal carcinoma: insights from germ-free and conventional animal models. Cancer J. 20, 217–224 (2014).
Marcobal, A. et al. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 7, 1933–1943 (2013).
Swann, J. et al. Gut microbiome modulates the toxicity of hydrazine: a metabonomic study. Mol. Biosyst. 5, 351–355 (2009).
Marchesi, J. R. et al. Towards the human colorectal cancer microbiome. PLoS ONE 6, e20447 (2011).
Magnusdottir, S., Ravcheev, D., de Crecy-Lagard, V. & Thiele, I. Systematic genome assessment of B-vitamin biosynthesis suggests co-operation among gut microbes. Front. Genet. 6, 148 (2015).
Raman, M. et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 11, 868–875 (2013).
Kelly, C. R. et al. Fecal microbiota transplant for treatment of Clostridium difficile infection in immunocompromised patients. Am. J. Gastroenterol. 109, 1065–1071 (2014).
Eiseman, B., Silen, W., Bascom, G. S. & Kauvar, A. J. Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis. Surgery 44, 854–859 (1958).
Gough, E., Shaikh, H. & Manges, A. R. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin. Infect. Dis. 53, 994–1002 (2011).
Weingarden, A. R. et al. Microbiota transplantation restores normal fecal bile acid composition in recurrent Clostridium difficile infection. Am. J. Physiol. Gastrointest. Liver Physiol. 306, G310–G319 (2014).
Borody, T. J., Brandt, L. J. & Paramsothy, S. Therapeutic faecal microbiota transplantation: current status and future developments. Curr. Opin. Gastroenterol. 30, 97–105 (2014).
Colman, R. J. & Rubin, D. T. Fecal microbiota transplantation as therapy for inflammatory bowel disease: A systematic review and meta-analysis. J. Crohns Colitis 8, 1569–1581 (2014).
Hanauer, S. B. & Sandborn, W. Management of Crohn's disease in adults. Am. J. Gastroenterol. 96, 635–643 (2001).
Seeley, E. H., Washington, M. K., Caprioli, R. M. & M'Koma, A. E. Proteomic patterns of colonic mucosal tissues delineate Crohn's colitis and ulcerative colitis. Proteomics Clin. Appl. 7, 541–549 (2013).
Maus, B. et al. Molecular reclassification of Crohn's disease: a cautionary note on population stratification. PLoS ONE 8, e77720 (2013).
Gerich, M. E. & McGovern, D. P. Towards personalized care in IBD. Nat. Rev. Gastroenterol. Hepatol. 11, 287–299 (2014).
Dawiskiba, T. et al. Serum and urine metabolomic fingerprinting in diagnostics of inflammatory bowel diseases. World J. Gastroenterol. 20, 163–174 (2014).
Williams, H. R. et al. Serum metabolic profiling in inflammatory bowel disease. Dig. Dis. Sci. 57, 2157–2165 (2012).
Fathi, F. et al. 1H NMR based metabolic profiling in Crohn's disease by random forest methodology. Magn. Reson. Chem. 52, 370–376 (2014).
Kostic, A. D., Xavier, R. J. & Gevers, D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146, 1489–1499 (2014).
Sartor, R. B. Microbial influences in inflammatory bowel diseases. Gastroenterology 134, 577–594 (2008).
Elliott, P. et al. Urinary metabolic signatures of human adiposity. Sci. Transl. Med. 7, 285ra62 (2015).
Shaukat, A. et al. Long-term mortality after screening for colorectal cancer. N. Engl. J. Med. 369, 1106–1114 (2013).
Mirnezami, R. et al. Chemical mapping of the colorectal cancer microenvironment via MALDI imaging mass spectrometry (MALDI-MSI) reveals novel cancer-associated field effects. Mol. Oncol. 8, 39–49 (2014).
Chan, E. C. Y. et al. Metabolic profiling of human colorectal cancer using high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy and gas chromatography mass spectrometry (GC/MS). J. Proteome Res. 8, 352–361 (2009).
Wang, H. et al. 1H NMR-based metabolic profiling of human rectal cancer tissue. Mol. Cancer 12, 121 (2013).
Zhu, J. et al. Colorectal cancer detection using targeted serum metabolic profiling. J. Proteome Res. 13, 4120–4130 (2014).
Montrose, D. C. et al. Metabolic profiling, a noninvasive approach for the detection of experimental colorectal neoplasia. Cancer Prev. Res. 5, 1358–1367 (2012).
Nishiumi, S. et al. A novel serum metabolomics-based diagnostic approach for colorectal cancer. PLoS ONE 7, e40459 (2012).
Ibanez, C. et al. CE/LC-MS multiplatform for broad metabolomic analysis of dietary polyphenols effect on colon cancer cells proliferation. Electrophoresis 33, 2328–2336 (2012).
Tan, B. et al. Metabonomics identifies serum metabolite markers of colorectal cancer. J. Proteome Res. 12, 3000–3009 (2013).
Cross, A. J. et al. A prospective study of serum metabolites and colorectal cancer risk. Cancer 120, 3049–3057 (2014).
Silva, C. L., Passos, M. & Camara, J. S. Investigation of urinary volatile organic metabolites as potential cancer biomarkers by solid-phase microextraction in combination with gas chromatography-mass spectrometry. Br. J. Cancer 105, 1894–1904 (2011).
Eisner, R., Greiner, R., Tso, V., Wang, H. & Fedorak, R. N. A machine-learned predictor of colonic polyps based on urinary metabolomics. BioMed Res. Int. 2013, 303982 (2013).
Goedert, J. J. et al. Fecal metabolomics: assay performance and association with colorectal cancer. Carcinogenesis 35, 2089–2096 (2014).
Phua, L. C. et al. Non-invasive fecal metabonomic detection of colorectal cancer. Cancer Biol. Ther. 15, 389–397 (2014).
Wedemeyer, H., Dore, G. J. & Ward, J. W. Estimates on HCV disease burden worldwide - filling the gaps. J. Viral Hepat. 22 Suppl 1, 1–5 (2015).
Trepo, C., Chan, H. L. & Lok, A. Hepatitis B virus infection. Lancet 384, 2053–2063 (2014).
Gomaa, A. I., Khan, S. A., Toledano, M. B., Waked, I. & Taylor-Robinson, S. D. Hepatocellular carcinoma: epidemiology, risk factors and pathogenesis. World J. Gastroenterol. 14, 4300–4308 (2008).
Wiessing, L. et al. Hepatitis C virus infection epidemiology among people who inject drugs in Europe: a systematic review of data for scaling up treatment and prevention. PLoS ONE 9, e103345 (2014).
Gomaa, A. I., Hashim, M. S. & Waked, I. Comparing staging systems for predicting prognosis and survival in patients with hepatocellular carcinoma in egypt. PLoS ONE 9, e90929 (2014).
Kohli, A., Shaffer, A., Sherman, A. & Kottilil, S. Treatment of Hepatitis C: A systematic review. JAMA 312, 631–640 (2014).
Ladep, N. G. & Taylor-Robinson, S. D. Management of liver disease in Nigeria. Clin. Med. 7, 439–441 (2007).
Beyoglu, D. & Idle, J. R. The metabolomic window into hepatobiliary disease. J. Hepatol. 59, 842–858 (2013).
Dumas, M. E., Kinross, J. & Nicholson, J. K. Metabolic phenotyping and systems biology approaches to understanding metabolic syndrome and fatty liver disease. Gastroenterology 146, 46–62 (2014).
Cassol, E. et al. Plasma metabolomics identifies lipid abnormalities linked to markers of inflammation, microbial translocation, and hepatic function in HIV patients receiving protease inhibitors. BMC Infect. Dis. 13, 203 (2013).
Zhang, A. H., Sun, H., Han, Y., Yan, G. L. & Wang, X. J. Urinary metabolic biomarker and pathway study of hepatitis B virus infected patients based on UPLC-MS system. PLoS ONE 8, e64381 (2013).
Ladep, N. G. et al. Discovery and validation of urinary metabotypes for the diagnosis of hepatocellular carcinoma in West Africans. Hepatology 60, 1291–1301 (2014).
Bosch, F. X., Ribes, J., Diaz, M. & Cleries, R. Primary liver cancer: worldwide incidence and trends. Gastroenterology 127, S5–S16 (2004).
Patel, M. et al. Hepatocellular carcinoma: diagnostics and screening. J. Eval. Clin. Pract. 18, 335–342 (2012).
Johnson, P. J. Role of alpha-fetoprotein in the diagnosis and management of hepatocellular carcinoma. J. Gastroenterol. Hepatol. 14, S32–S36 (1999).
Farinati, F. et al. Diagnostic and prognostic role of alpha-fetoprotein in hepatocellular carcinoma: Both or neither? Am. J. Gastroenterol. 101, 524–532 (2006).
Bedossa, P. & Poynard, T. An algorithm for the grading of activity in chronic hepatitis, C. The METAVIR cooperative study group. Hepatology 24, 289–293 (1996).
Ishak, K. et al. Histological grading and staging of chronic hepatitis. J. Hepatol. 22, 696–699 (1995).
Baranova, A., Lal, P., Birerdinc, A. & Younossi, Z. M. Non-invasive markers for hepatic fibrosis. BMC Gastroenterol. 11, 91 (2011).
Regev, A. et al. Sampling error and intraobserver variation in liver biopsy in patients with chronic HCV infection. Am. J. Gastroenterol. 97, 2614–2618 (2002).
de Groen, P. C., Gores, G. J., LaRusso, N. F., Gunderson, L. L. & Nagorney, D. M. Biliary tract cancers. N. Engl. J. Med. 341, 1368–1378 (1999).
Zabron, A., Edwards, R. J. & Khan, S. A. The challenge of cholangiocarcinoma: Dissecting the molecular mechanisms of an insidious cancer. Dis. Model. Mech. 6, 281–292 (2013).
Sithithaworn, P., Yongvanit, P., Duenngai, K., Kiatsopit, N. & Pairojkul, C. Roles of liver fluke infection as risk factor for cholangiocarcinoma. J. Hepatobiliary Pancreat. Sci. 21, 301–308 (2014).
Khan, S. A., Thomas, H. C., Davidson, B. R. & Taylor-Robinson, S. D. Cholangiocarcinoma. Lancet 366, 1303–1314 (2005).
Shariff, M. I. et al. Characterization of urinary biomarkers of hepatocellular carcinoma using magnetic resonance spectroscopy in a Nigerian population. J. Proteome Res. 9, 1096–1103 (2010).
Canbakan, B. et al. Clinical, biochemical and histological correlations in a group of non-drinker subjects with non-alcoholic fatty liver disease. Acta Gastroenterol. Belg. 70, 277–284 (2007).
Dyson, J. K., Anstee, Q. M. & McPherson, S. Non-alcoholic fatty liver disease: a practical approach to diagnosis and staging. Frontline Gastroenterol. 5, 211–218 (2014).
LaBrecque, D. R. et al. World Gastroenterology Organisation global guidelines: Nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. J. Clin. Gastroenterol. 48, 467–473 (2014).
Corey, K. E. & Kaplan, L. M. Obesity and liver disease: The epidemic of the twenty-first century. Clin. Liver Dis. 18, 1–18 (2014).
Gomaa, A. I., Khan, S. A., Leen, E. L., Waked, I. & Taylor-Robinson, S. D. Diagnosis of hepatocellular carcinoma. World J. Gastroenterol. 15, 1301–1314 (2009).
Barr, J. et al. Liquid chromatography-mass spectrometry-based parallel metabolic profiling of human and mouse model serum reveals putative biomarkers associated with the progression of nonalcoholic fatty liver disease. J. Proteome Res. 9, 4501–4512 (2010).
Barr, J. et al. Obesity-dependent metabolic signatures associated with nonalcoholic fatty liver disease progression. J. Proteome Res. 11, 2521–2532 (2012).
Clayton, T. A., Baker, D., Lindon, J. C., Everett, J. R. & Nicholson, J. K. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl Acad. Sci. USA 106, 14728–14733 (2009).
Wang, Z. et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472, 57–63 (2011).
Yap, I. K. et al. Metabolome-wide association study identifies multiple biomarkers that discriminate north and south Chinese populations at differing risks of cardiovascular disease: INTERMAP study. J. Proteome Res. 9, 6647–6654 (2010).
Arteaga, C. L. et al. Treatment of HER2-positive breast cancer: Current status and future perspectives. Nat. Rev. Clin. Onc. 9, 16–32 (2012).
Gutiu, I. A. et al. Pharmacometabonomics, pharmacogenomics and personalized medicine. Rom. J. Intern. Med. 48, 187–191 (2010).
Kwon, H. N. et al. Predicting idiopathic toxicity of cisplatin by a pharmacometabonomic approach. Kidney Int. 79, 529–537 (2011).
Nicholson, J. K., Wilson, I. D. & Lindon, J. C. Pharmacometabonomics as an effector for personalized medicine. Pharmacogenomics 12, 103–111 (2011).
Backshall, A., Sharma, R., Clarke, S. J. & Keun, H. C. Pharmacometabonomic profiling as a predictor of toxicity in patients with inoperable colorectal cancer treated with capecitabine. Clin. Cancer Res. 17, 3019–3028 (2011).
Everett, J. R., Loo, R. L. & Pullen, F. S. Pharmacometabonomics and personalized medicine. Ann. Clin. Biochem. 50, 523–545 (2013).
K. C. et al. In vivo, in situ tissue analysis using rapid evaporative ionization mass spectrometry. Angew. Chem. Int. Ed. Engl. 48, 8240–8242 (2009).
Balog, J. et al. Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Sci. Transl. Med. 5, 194ra93 (2013).
Strittmatter, N. et al. Analysis of intact bacteria using rapid evaporative ionisation mass spectrometry. Chem. commun. 49, 6188–6190 (2013).
Balog, J. et al. Identification of biological tissues by rapid evaporative ionization mass spectrometry. Anal. Chem. 82, 7343–7350 (2010).
Plichta, J. K. et al. Does practice make perfect? Resident experience with breast surgery influences excision adequacy. Am. J. Surg. 209, 547–551 (2015).
Guenther, S. et al. Spatially resolved metabolic phenotyping of breast cancer by desorption electrospray ionization mass spectrometry. Cancer Res. 75, 1828–1837 (2015).
Nicholson, J. K. & Lindon, J. C. Systems biology: Metabonomics. Nature 455, 1054–1056 (2008).
Lindon, J. C. et al. Summary recommendations for standardization and reporting of metabolic analyses. Nat. Biotechnol. 23, 833–838 (2005).
MRC-NIHR National Phenome Centre [online]
Dona, A. C. et al. Precision high-throughput proton NMR spectroscopy of human urine, serum, and plasma for large-scale metabolic phenotyping. Anal. Chem. 86, 9887–9894 (2014).
Sarafian, M. H. et al. Objective set of criteria for optimization of sample preparation procedures for ultra-high throughput untargeted blood plasma lipid profiling by ultra performance liquid chromatography-mass spectrometry. Anal. Chem. 86, 5766–5774 (2014).
Human Metabolome Database [online]
Mirnezami, R. et al. Rapid diagnosis and staging of colorectal cancer via high-resolution magic angle spinning nuclear magnetic resonance (HR-MAS NMR) spectroscopy of intact tissue biopsies. Ann. Surg. 259, 1138–1149 (2014).
Cajka, T. & Fiehn, O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. Trends Analyt. Chem. 61, 192–206 (2014).
Kurland, I. J. et al. Application of combined omics platforms to accelerate biomedical discovery in diabesity. Ann. N. Y. Acad. Sci. 1287, 1–16 (2013).
Nicholson, J. K., Holmes, E., Lindon, J. C. & Wilson, I. D. The challenges of modeling mammalian biocomplexity. Nat. Biotechnol. 22, 1268–1274 (2004).
Biological Magnetic Resonance Data Bank [online]
Acknowledgements
The MRC-NIHR National Phenome Centre is supported by the UK Medical Research Council (in association with National Institute for Health Research [England]) Grant MC_PC_12025. The financial support of Bruker Biospin, Waters Corporation, Metabometrix and Imperial College is also gratefully acknowledged by the MRC-NIHR National Phenome Centre.
Author information
Authors and Affiliations
Contributions
E.H., A.W., S.D.T.-R. and J.K.N. researched data for and wrote the article. E.H., A.W. and J.K.N. reviewed/edited the manuscript before submission and E.H. and J.K.N. made substantial contributions to discussion of content.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Table 1
Analytical techniques for metabolic phenotyping and examples of clinical applications (PDF 123 kb)
Rights and permissions
About this article
Cite this article
Holmes, E., Wijeyesekera, A., Taylor-Robinson, S. et al. The promise of metabolic phenotyping in gastroenterology and hepatology. Nat Rev Gastroenterol Hepatol 12, 458–471 (2015). https://doi.org/10.1038/nrgastro.2015.114
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrgastro.2015.114
This article is cited by
-
Identifying disease-related microbes based on multi-scale variational graph autoencoder embedding Wasserstein distance
BMC Biology (2023)
-
Diagnosis and Management of Noncirrhotic Portal Hypertension
Current Hepatology Reports (2023)
-
Metabolomics and health: from nutritional crops and plant-based pharmaceuticals to profiling of human biofluids
Cellular and Molecular Life Sciences (2021)
-
WMGHMDA: a novel weighted meta-graph-based model for predicting human microbe-disease association on heterogeneous information network
BMC Bioinformatics (2019)
-
Tsumura-Suzuki obese diabetic mice-derived hepatic tumors closely resemble human hepatocellular carcinomas in metabolism-related genes expression and bile acid accumulation
Hepatology International (2018)