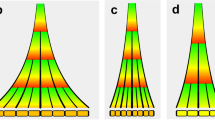
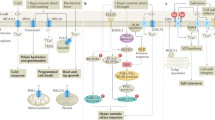
Key Points
-
Indeterminate growth is a unique feature of plant development that is subject to intrinsic controls and modulated by environmental cues.
-
Growth control is mediated by hormonal responses that rapidly alter gene expression programmes by inducing or preventing degradation of transcriptional regulators by the ubiquitin–proteasome system.
-
Developmental targets of hormones are the meristems (stem cells and their transit amplifying daughter cells), lateral organ founder cells and the developing organ primordia.
-
Plant development is modulated by the well-known classic hormones, such as auxin, cytokinin and gibberellin, and by newly identified growth regulators, such as peptides, strigolactone and fatty acid-related molecules. Numerous growth regulators are yet to be discovered.
-
Hormones do not act in isolation but are interrelated by synergistic or antagonistic crosstalk so that the hormones modulate each other's biosynthesis or response.
-
Hormones stimulate or terminate cell proliferation and thus regulate meristem or organ size, depending on the spatial distribution of their receptors and transcriptional regulators, as well as the relative concentrations of antagonistically acting hormones such as auxin and cytokinin.
-
Environmental cues, such as light or stress, elicit stimulatory or inhibitory growth changes by altering local hormone biosynthesis or response.
-
Despite tremendous recent progress, important aspects of hormone action in development are yet to be analysed, including hormone-regulated cell fate and tissue-specific hormone responses.
Abstract
Plant development is subject to hormonal growth control and adapts to environmental cues such as light or stress. Recently, significant progress has been made in elucidating hormone synthesis, signalling and degradation pathways, and in resolving spatial and temporal aspects of hormone responses. Here we review how hormones control maintenance of stem cell systems, influence developmental transitions of stem cell daughters and define developmental compartments in Arabidopsis thaliana. We also discuss how environmental cues change plant growth by modulating hormone levels and response. Future analysis of hormone crosstalk and of hormone action at both single cell and organ levels will substantially improve our understanding of how plant development adapts to changes in intrinsic and environmental conditions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Benková, E. & Hejátko, J. Hormone interactions at the root apical meristem. Plant Mol. Biol. 69, 383–396 (2008).
Dugardeyn, J., Vandenbussche, F. & Van Der Straeten, D. To grow or not to grow: what can we learn on ethylene–gibberellin cross-talk by in silico gene expression analysis? J. Exp. Bot. 59, 1–16 (2008).
Hirose, N. et al. Regulation of cytokinin biosynthesis, compartmentalization and translocation. J. Exp. Bot. 59, 75–83 (2008).
Bishopp, A. et al. Signs of change: hormone receptors that regulate plant development. Development 133, 1857–1869 (2006).
Dharmasiri, N. et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 9, 109–119 (2005).
Thines, B. et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665 (2007).
Chini, A. et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671 (2007).
Dreher, K. & Callis, J. Ubiquitin, hormones and biotic stress in plants. Ann. Bot. 99, 787–822 (2007).
Schwechheimer, C., Serino, G. & Deng, X. W. Multiple ubiquitin ligase-mediated processes require COP9 signalosome and AXR1 function. Plant Cell 14, 2553–2563 (2002).
Higuchi, M. et al. In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. USA 101, 8821–8826 (2004).
Nishimura, C. et al. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 16, 1365–1377 (2004).
Cheng, Y., Dai, X. & Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799 (2006).
Cheng, Y., Dai, X. & Zhao, Y. Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19, 2430–2439 (2007).
Brady, S. M. et al. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318, 801–806 (2007).
Dinneny, J. R. et al. Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320, 942–945 (2008).
Savaldi-Goldstein, S., Peto, C. & Chory, J. The epidermis both drives and restricts plant shoot growth. Nature 446, 199–202 (2007).
Dello Ioio, R. et al. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17, 678–682 (2007).
Ubeda-Tomás, S. et al. Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nature Cell Biol. 10, 625–628 (2008).
Galinha, C. et al. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449, 1053–1057 (2007). This is the first paper to describe a protein gradient in plants. PLT protein levels were linked to concentration-dependent PLT responses in different root developmental zones.
Swarup, R. et al. Root gravitropism requires lateral root cap and epidermal cells for transport and response to a mobile auxin signal. Nature Cell Biol. 7, 1057–1065 (2005).
Jenik, P. D., Gillmor, C. S. & Lukowitz, W. Embryonic patterning in Arabidopsis thaliana. Annu. Rev. Cell Dev. Biol. 23, 207–236 (2007).
Friml, J. et al. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 426, 147–153 (2003).
Aida, M. et al. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120 (2004).
Weijers, D. et al. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev. Cell 10, 265–270 (2006).
Müller, B. & Sheen, J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature 453, 1094–1097 (2008).
Riefler, M., Novak, O., Strnad, M. & Schmülling, T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 18, 40–54 (2006).
Long, J. A., Ohno, C., Smith, Z. R. & Meyerowitz, E. M. TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312, 1520–1523 (2006).
Szemenyei, H., Hannon, M. & Long, J. A. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386 (2008).
Treml, B. S. et al. The gene ENHANCER OF PINOID controls cotyledon development in the Arabidopsis embryo. Development 132, 4063–4074 (2005).
Breuninger, H., Rikirsch, E., Hermann, M., Ueda, M. & Laux, T. Differential expression of WOX genes mediates apical-basal axis formation in the Arabidopsis embryo. Dev. Cell 14, 867–876 (2008).
Furutani, M. et al. PIN-FORMED1 and PINOID regulate boundary formation and cotyledon development in Arabidopsis embryogenesis. Development 131, 5021–5030 (2004).
Mayer, K. F. et al. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95, 805–815 (1998).
Long, J. A., Moan, E. I., Medford, J. I. & Barton, M. K. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69 (1996).
De Smet, I. et al. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134, 681–690 (2007).
Dello Ioio, R. et al. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322, 1380–1384 (2008). This paper identifies the CK and auxin signalling components that are required to set up the root meristem boundary and resolves the basis for auxin–CK antagonism in the root.
Fu, X. & Harberd, N. P. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421, 740–743 (2003). The DELLA repressors are identified as fundamental regulators of auxin-mediated root growth responses.
Lenhard, M., Jürgens, G. & Laux, T. The WUSCHEL and SHOOTMERISTEMLESS genes fulfil complementary roles in Arabidopsis shoot meristem regulation. Development 129, 3195–3206 (2002).
Schoof, H. et al. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644 (2000).
Scheres, B. Stem-cell niches: nursery rhymes across kingdoms. Nature Rev. Mol. Cell Biol. 8, 345–354 (2007).
Jasinski, S. et al. KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15, 1560–1565 (2005). KNOX genes are identified as central regulators of CK and GA hormone homeostasis by oppositely affecting CK and GA levels in the shoot meristem.
Yanai, O. et al. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr. Biol. 15, 1566–1571 (2005).
Hay, A. et al. The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 12, 1557–1565 (2002). The authors describe how KNOX-mediated meristematic maintenance in the shoot meristem and in leaves is antagonized by gibberellin.
Hamant, O. et al. The KNAT2 homeodomain protein interacts with ethylene and cytokinin signaling. Plant Physiol. 130, 657–665 (2002).
Leibfried, A. et al. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438, 1172–1175 (2005).
Kyozuka, J. Control of shoot and root meristem function by cytokinin. Curr. Opin. Plant Biol. 10, 442–446 (2007).
Reinhardt, D. et al. Regulation of phyllotaxis by polar auxin transport. Nature 426, 255–260 (2003).
Nordström, A. et al. Auxin regulation of cytokinin biosynthesis in Arabidopsis thaliana: a factor of potential importance for auxin–cytokinin-regulated development. Proc. Natl. Acad. Sci. USA 101, 8039–8044 (2004).
Heisler, M. G. et al. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15, 1899–1911 (2005).
Bainbridge, K. et al. Auxin influx carriers stabilize phyllotactic patterning. Genes Dev. 22, 810–823 (2008).
Shani, E., Yanai, O. & Ori, N. The role of hormones in shoot apical meristem function. Curr. Opin. Plant Biol. 9, 484–489 (2006).
Weiss, D. & Ori, N. Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol. 144, 1240–1246 (2007).
Fiers, M., Ku, K. L. & Liu, C. M. CLE peptide ligands and their roles in establishing meristems. Curr. Opin. Plant Biol. 10, 39–43 (2007).
Kepinski, S. Integrating hormone signaling and patterning mechanisms in plant development. Curr. Opin. Plant Biol. 9, 28–34 (2006).
Blilou, I. et al. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433, 39–44 (2005).
Grieneisen, V. A., Xu, J., Marée, A. F., Hogeweg, P. & Scheres, B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449, 1008–1013 (2007).
Stepanova, A. N., Hoyt, J. M., Hamilton, A. A. & Alonso, J. M. A Link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17, 2230–2242 (2005).
Stepanova, A. N. et al. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191 (2008). The TAA1/TAR family of auxin synthesis genes are shown to be as crucial for embryonic and postembryonic development, and the authors reveal direct regulation of auxin synthesis as a central element of ethylene-modulated growth.
Miyawaki, K., Matsumoto-Kitano, M. & Kakimoto, T. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 37, 128–138 (2004).
Fukuda, H. Signals that control plant vascular cell differentiation. Nature Rev. Mol. Cell Biol. 5, 379–391 (2004).
Carabelli, M. et al. Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev. 21, 1863–1868 (2007). This paper links differential regulation of organ growth to auxin-mediated CK breakdown and implicates auxin synthesis as a primary tool for light-induced growth promotion (by a low ratio of red to far-red light), as well as growth restriction.
Werner, T., Köllmer, I., Bartrina, I., Holst, K. & Schmülling, T. New insights into the biology of cytokinin degradation. Plant Biol. 8, 371–381 (2006).
Frigerio, M. et al. Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol. 142, 553–563 (2006).
Nemhauser, J. L., Hong, F. & Chory, J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126, 467–475 (2006).
Silverstone, A. L. et al. Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J. 12, 9–19 (1997).
Mitchum, M. G. et al. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 45, 804–818 (2006).
Brenner, W. G., Romanov, G. A., Köllmer, I., Bürkle, L. & Schmülling, T. Immediate–early and delayed cytokinin response genes of Arabidopsis thaliana identified by genome-wide expression profiling reveal novel cytokinin-sensitive processes and suggest cytokinin action through transcriptional cascades. Plant J. 44, 314–333 (2005).
Shimada, A. et al. The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J. 48, 390–402 (2006).
Greenboim-Wainberg, Y. et al. Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell 17, 92–102 (2005).
Swain, S. M. et al. Altered expression of SPINDLY affects gibberellin response and plant development. Plant Phys. 126, 1174–1185 (2001).
Sarkar, A. K. et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446, 811–814 (2007).
Kondo, T. et al. A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313, 845–848 (2006).
De Veylder, L., Beeckman, T. & Inze, D. The ins and outs of the plant cell cycle. Nature Rev. Mol. Cell Biol. 8, 655–665 (2007).
del Pozo, J. C., Boniotti, M. B. & Gutierrez, C. Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(AtSKP2) pathway in response to light. Plant Cell 14, 3057–3071 (2002).
Riou-Khamlichi, C., Menges, M., Healy, J. M. & Murray, J. A. Sugar control of the plant cell cycle: differential regulation of Arabidopsis D-type cyclin gene expression. Mol. Cell. Biol. 20, 4513–4521 (2000).
Tao, Y. et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176 (2008). This paper shows that the auxin biosynthesis gene TAA1 is regulated by a low ratio of red to far-red light to rapidly increase the level of auxin required for the shade avoidance response.
Scarpella, E., Marcos, D., Friml, J. & Berleth, T. Control of leaf vascular patterning by polar auxin transport. Genes Dev. 20, 1015–1027 (2006).
Anastasiou, E. et al. Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev. Cell 13, 843–856 (2007). A novel, as yet unidentified, KLUH-derived signal is implicated in orchestrating aerial organ growth.
Mizukami, Y. & Fischer, R. L. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proc. Natl. Acad. Sci. USA 97, 2942–2947 (2000).
Nole-Wilson, S., Tranby, T. L. & Krizek, B. A. AINTEGUMENTA-like (AIL) genes are expressed in young tissues and may specify meristematic or division-competent states. Plant Mol. Biol. 57, 613–628 (2005).
Mizukami, Y. A matter of size: developmental control of organ size in plants. Curr. Opin. Plant Biol. 4, 533–539 (2001).
Hu, Y., Xie, Q. & Chua, N. H. The Arabidopsis auxin-inducible gene ARGOS controls lateral organ size. Plant Cell 15, 1951–1961 (2003).
Werner, T. et al. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15, 2532–2550 (2003).
Rashotte, A. M. et al. A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci. USA 103, 11081–11085 (2006).
Disch, S. et al. The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 16, 272–279 (2006).
Fleet, C. M. & Sun, T. A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 8, 77–85 (2005).
Teale, W. D., Paponov, I. A. & Palme, K. Auxin in action: signalling, transport and the control of plant growth and development. Nature Rev. Mol. Cell Biol. 7, 847–859 (2006).
Hu, Y., Poh, H. M. & Chua, N. H. The Arabidopsis ARGOS-LIKE gene regulates cell expansion during organ growth. Plant J. 47, 1–9 (2006).
Nakaya, M., Tsukaya, H., Murakami, N. & Kato, M. Brassinosteroids control the proliferation of leaf cells of Arabidopsis thaliana. Plant Cell Physiol. 43, 239–244 (2002).
Ogas, J. et al. Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277, 91–94 (1997).
Henderson, J. T. et al. PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Phys. 134, 995–1005 (2004).
Hirakawa, Y. et al. Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 105, 15208–15213 (2008).
Whitford, R. et al. Plant CLE peptides from two distinct functional classes synergistically induce division of vascular cells. Proc. Natl. Acad. Sci. USA 105, 18625–18630 (2008).
Vidaurre, D. P., Ploense, S., Krogan, N. T. & Berleth, T. AMP1 and MP antagonistically regulate embryo and meristem development in Arabidopsis. Development 134, 2561–2567 (2007).
Gomez-Roldan, V. et al. Strigolactone inhibition of shoot branching. Nature 455, 189–194 (2008).
Umehara, M. et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200 (2008).
Rolland-Lagan, A., Bangham, J. A. & Coen, E. Growth dynamics underlying petal shape and asymmetry. Nature 422, 161–163 (2003).
Reinhardt, B. et al. Restoration of DWF4 expression to the leaf margin of a dwf4 mutant is sufficient to restore leaf shape but not size: the role of the margin in leaf development. Plant J. 52, 1094–1104 (2007).
Achard, P. et al. Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94 (2006). This paper describes the central role of the DELLA repressors in the regulation of overall plant growth in response to stresses.
Spoel, S. H. & Dong, X. Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3, 348–351 (2008).
Bhalerao, R. P. et al.Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J. 29, 325–332 (2002).
Park, J. E. et al. GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. J. Biol. Chem. 282, 10036–10046 (2007).
Wang, D., Pajerowska-Mukhtar, K., Culler, A. H. & Dong, X. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr. Biol. 17, 1784–1790 (2007).
Huang, D., Wu, W., Abrams, S. R. & Cutler, A. J. The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 59, 2991–3007 (2008).
Navarro, L. et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439 (2006).
Achard, P., Vriezen, W. H., Van Der Straeten, D. & Harberd, N. P. Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15, 2816–2825 (2003).
Pérez-Torres, CA. et al. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell 20, 3258–3272 (2008).
Stuttmann, J. et al. COP9 signalosome- and 26S Proteasome-dependent regulation of SCFTIR1 accumulation in Arabidopsis. J. Biol. Chem. 284, 7920–7930 (2009).
Malamy, J. E. Intrinsic and environmental response pathways that regulate root system architecture. Plant Cell Environ. 28, 67–77 (2005).
Walch-Liu, P. et al. Nitrogen regulation of root branching. Ann. Bot. (Lond.) 97, 875–881 (2006).
Jiao, Y., Lau, O. S. & Deng, X. W. Light-regulated transcriptional networks in higher plants. Nature Rev. Genet. 8, 217–230 (2007).
Oh, E. et al. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19, 1192–1208 (2007).
de Lucas, M. et al. A molecular framework for light and gibberellin control of cell elongation. Nature 451, 480–484 (2008).
Feng, S. et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451, 475–479 (2008).
Devlin, P. F., Yanovsky, M. J. & Kay, S. A. A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol. 133, 1617–1629 (2003).
Kanyuka, K. et al. Mutations in the huge Arabidopsis gene BIG affect a range of hormone and light responses. Plant J. 35, 57–70 (2003).
Nambara, E. & Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 56, 165–185 (2005).
Broekaert, W. F., Delauré, S. L., De Bolle, M. F. & Cammue, B. P. The role of ethylene in host–pathogen interactions. Annu. Rev. Phytopathol. 44, 393–416 (2006).
Alonso, J. M., Hirayama, T., Roman, G., Nourizadeh, S. & Ecker, J. R. EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284, 2148–2152 (1999).
León, P. & Sheen, J. Sugar and hormone connections. Trends Plant Sci. 8, 110–116 (2003).
Cheng, WH. et al. A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14, 2723–2743 (2002).
Srivastava, R., Liu, J. X. & Howell, S. H. Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilisin serine protease in Arabidopsis. Plant J. 56, 219–227 (2008).
Stenvik, G. E. et al. The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20, 1805–1817 (2008).
Benková, E. et al. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602 (2003).
Blakeslee, J. J. et al. Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 19, 131–147 (2007).
Woodward, A. W. & Bartel, B. Auxin: regulation, action, and interaction. Ann. Bot. 95, 707–735 (2005).
Kurakawa, T. et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655 (2007).
Yamaguchi, S. Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251 (2008).
Schwechheimer, C. Understanding gibberellic acid signaling — are we there yet? Curr. Opin. Plant Biol. 11, 9–15 (2008).
Murase, K., Hirano, Y., Sun, T. P. & Hakoshima, T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463 (2008).
Geldner, N., Hyman, D. L., Wang, X., Schumacher, K. & Chory, J. Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 21, 1598–1602 (2007).
Symons, G. M., Ross, J. J., Jager, C. E. & Reid, J. B. Brassinosteroid transport. J. Exp. Bot. 59, 17–24 (2008).
Ohnishi, T. et al. Tomato cytochrome P450 CYP734A7 functions in brassinosteroid catabolism. Phytochem. 67, 1895–1906 (2006).
Wasternack, C. Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100, 681–697 (2007).
Lorenzo, O., Chico, J. M., Sánchez-Serrano, J. J. & Solano, R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16, 1938–1950 (2004).
McCourt, P. & Creelman, R. The ABA receptors — we report you decide. Curr. Opin. Plant Biol. 11, 474–478 (2008).
Jiang, F. & Hartung, W. Long-distance signalling of abscisic acid (ABA): the factors regulating the intensity of the ABA signal. J. Exp. Bot. 59, 37–43 (2008).
Kendrick, M. D. & Chang, C. Ethylene signaling: new levels of complexity and regulation. Curr. Opin. Plant Biol. 11, 479–485 (2008).
Hartig, K. & Beck, E. Crosstalk between auxin, cytokinins, and sugars in the plant cell cycle. Plant Biol. 8, 389–396 (2006).
Magyar, Z. et al. The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17, 2527–2541 (2005).
Dewitte, W. & Murray, J. A. The plant cell cycle. Annu. Rev. Plant Biol. 54, 235–264 (2003).
Vanneste, S. et al. Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17, 3035–3050 (2005).
Achard, P. et al. DELLAs contribute to plant photomorphogenesis. Plant Physiol. 143, 1163–1172 (2007).
Sullivan, J. A., Shirasu, K. & Deng, X. W. The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nature Rev. Genet. 4, 948–958 (2003).
To, J. P. & Kieber, J. J. Cytokinin signaling: two-components and more. Trends Plant Sci. 13, 85–92 (2008).
Acknowledgements
We would like to thank C. Kägi for help with the figures and for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Related links
Glossary
- F-box receptor
-
The component of an SCF E3 ubiquitin ligase that acts as a hormone receptor and that recognizes repressor proteins as substrates to be targeted for degradation.
- Meristem
-
A plant stem cell system comprising the niche, the pluripotent stem cells and their highly proliferating daughter cells.
- Determinate organ
-
Plant organ that does not have unlimited growth potential but that differentiates once the final size is reached, for example, a leaf.
- Quiescent centre
-
A group of cells located in the centre of the root meristem and required to keep the surrounding stem cells from differentiating.
- Histone acetyltransferase
-
An enzyme that adds acetyl groups to lysine residues in the DNA-binding histone group of proteins and thereby modulates the transcriptional activity of genes.
- Chromatin remodelling
-
Transient changes in chromatin accessibility.
- Cotyledon
-
The leaves of a seedling that are formed during embryonic development.
- Indeterminate
-
Refers to a pattern of growth and development that is open ended. In meristems, this is associated with the continuous ability to produce determinate lateral organs, such as leaves. By contrast, determinate refers to growth and development that is restricted in time or space.
- Organizing centre
-
A group of cells that is located in the centre of the shoot meristem and is required to prevent the surrounding stem cells from differentiating.
- Transit amplifying cells
-
Stem cell daughters that divide for a definite number of times before they leave the meristem to undergo terminal differentiation.
- Transition zone
-
Boundary between the meristematic zone (which is proliferative) and the differentiation zones; it is easily recognized by the onset of cell elongation in the root.
- DELLA proteins
-
DELLA domain-containing gibberellin signalling repressors, which are named after a conserved stretch of amino acids that are required for their degradation by the 26S proteasome.
- CLE family
-
Family of CLV3-related peptide ligands that bind to receptor kinases that contain leucine-rich repeats, such as CLV1.
- Hormonal regime
-
Describes a situation in which the hormone concentration or the concentration ratio of different hormones mediates cell- or compartment-specific responses.
- Callus
-
A disorganized group of proliferating or undifferentiated cells.
- Lateral organ
-
An organ that is produced from the shoot apical meristem. Includes leaves and putatively homologous organs such as cotyledons (embryonic leaves), bracts (modified leaves that subtend reproductive structures) and floral organs.
- Signalosome
-
A multifunctional protein complex essential for development and possibly involved in the regulation of protein degradation.
- Proteasome
-
A large intracellular protein complex that degrades soluble proteins that have been modified by ubiquitylation.
- Photomorphogenesis
-
A light-dependent developmental process, such as seed germination or seedling growth.
- Phytochrome
-
One of three classes of known plant photoreceptors. Phytochromes are composed of a protein moiety covalently associated with a tetrapyrrole chromophore; they are synthesized in a red-light-absorbing form and are converted by red light to a far-red-light-absorbing isoforms, which bind to nuclear-localized transcription factors of the PIF class.
- Hypocotyl
-
The stem of a seedling.
Rights and permissions
About this article
Cite this article
Wolters, H., Jürgens, G. Survival of the flexible: hormonal growth control and adaptation in plant development. Nat Rev Genet 10, 305–317 (2009). https://doi.org/10.1038/nrg2558
Issue Date:
DOI: https://doi.org/10.1038/nrg2558
This article is cited by
-
Ethylene may be the Key Factor Leading to the Homologous Transformation of Stamens into Pistils in Three-Pistil Wheat
Journal of Plant Growth Regulation (2024)
-
A GARP transcription factor SlGCC positively regulates lateral root development in tomato via auxin-ethylene interplay
Planta (2024)
-
Oxygenation alleviates waterlogging-caused damages to cherry rootstocks
Molecular Horticulture (2023)
-
Efficient strategies for controlled release of nanoencapsulated phytohormones to improve plant stress tolerance
Plant Methods (2023)
-
Season, storage and extraction method impact on the phytochemical profile of Terminalia ivorensis
BMC Plant Biology (2023)