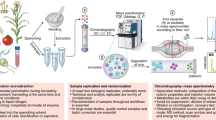
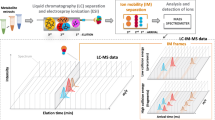
Abstract
Metabolome analysis by flow injection electrospray mass spectrometry (FIE-MS) fingerprinting generates measurements relating to large numbers of m/z signals. Such data sets often exhibit high variance with a paucity of replicates, thus providing a challenge for data mining. We describe data preprocessing and modeling methods that have proved reliable in projects involving samples from a range of organisms. The protocols interact with software resources specifically for metabolomics provided in a Web-accessible data analysis package FIEmspro (http://users.aber.ac.uk/jhd) written in the R environment and requiring a moderate knowledge of R command-line usage. Specific emphasis is placed on describing the outcome of modeling experiments using FIE-MS data that require further preprocessing to improve quality. The salient features of both poor and robust (i.e., highly generalizable) multivariate models are outlined together with advice on validating classifiers and avoiding false discovery when seeking explanatory variables.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout












Similar content being viewed by others
References
Somorjai, R.L., Dolenko, B. & Baumgartner, R. Class prediction and discovery using gene microarray and proteomics mass spectroscopy data: curses, caveats, cautions. Bioinformatics 19, 1484–1491 (2003).
Berrar, D., Bradbury, I. & Dubitzky, W. Avoiding model selection bias in small-sample genomic datasets. Bioinformatics 22, 1245–50 (2006).
BragaNeto, U.M. & Dougherty, E.R. Is cross-validation valid for small-sample microarray classification? Bioinformatics 20, 374–380 (2004).
Lyons-Weiler, J. et al. Assessing the statistical significance of the achieved classification error of classifiers constructed using serum peptide profiles, and a prescription for random sampling repeated studies for massive high-throughput genomic and proteomic studies. Cancer Inform. 1, 53–77 (2005).
Broadhurst, D.I. & Kell, D.B. Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics 2, 171–196 (2006).
Saghatelian, A. & Cravatt, B.F. Global strategies to integrate the proteome and metabolome. Curr. Opin. Chem. Biol. 9, 62–68 (2005).
EinDor, L., Zuk, O. & Domany, E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl. Acad. Sci. USA 103, 5923–5928 (2006).
Dyaz-Uriarte, R. Supervised methods with genomic data: a review and cautionary view. Data Analysis and Visualization in Genomics and Proteomics. pp 193–214 Wiley, New York, (2005).
Fawcett, T. ROC Graphs: Notes and Practical Considerations for Data Mining Researchers. Technical report HPL-2003-4. HP Laboratories, Palo Alto, CA, Available at http://www.hpl.hp.com/techreports/2003/HPL-2003-4.pdf (2003).
Mukherjee, S., Roberts, S.J. & van der Laan, M.J. Data-adaptive test statistics for microarray data. Bioinformatics 21, 108–114 (2005).
Sima, C. & Dougherty, E.R. What should be expected from feature selection in small-sample settings. Bioinformatics 22, 2430–2436 (2006).
Enot, D.P., Beckmann, M., Overy, D. & Draper, J. Predicting interpretability of metabolome models based on behavior, putative identity, and biological relevance of explanatory signals. Proc. Natl. Acad. Sci. USA 103, 14865–14870 (2006).
Kell, D.B., Darby, R.M. & Draper, J. Genomic computing. Explanatory analysis of plant expression profiling data using machine learning. Plant Physiol. 126, 943–951 (2001).
Catchpole, G.S. et al. Hierarchical metabolomics demonstrates substantial compositional similarity between genetically modified and conventional potato crops. Proc. Natl. Acad. Sci. USA 102, 14458–14462 (2005).
Goodacre, R., Vaidyanathan, S., Dunn, W.B., Harrigan, G.G. & Kell, D.B. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 22, 245–252 (2004).
Bino, R.J. et al. Potential of metabolomics as a functional genomics tool. Trends Plant Sci. 9, 418–425 (2004).
Fiehn, O. et al. Metabolite profiling for plant functional genomics. Nat. Biotechnol. 18, 1157–1161 (2000).
Sumner, L.W., Mendes, P. & Dixon, R.A. Plant metabolomics: large-scale phytochemistry in the functional genomics era. Phytochemistry 62, 817–836 (2003).
Nicholson, J.K. & Wilson, I.D. Understanding 'global' systems biology: metabonomics and the continuum of metabolism. Nat. Rev. Drug Discov. 2, 668–676 (2003).
Roessner, U., Wagner, C., Kopka, J., Trethewey, R.N. & Willmitzer, L. Simultaneous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J 23, 131–142 (2000).
Tolstikov, V.V. & Fiehn, O. Analysis of highly polar compounds of plant origin: Combination of hydrophilic interaction chromatography and electrospray ion trap mass spectrometry. Anal. Biochem. 301, 298–307 (2002).
Beckmann, M., Enot, D.P., Overy, D.P. & Draper, J. Representation, comparison and interpretation of metabolome fingerprint data for total composition analysis and quality trait investigation in potato cultivars. J. Agricultural and Food Chemistry 55, 3444–3451 (2007).
Dear, G.J., James, A.D. & Sarda, S. Ultra-performance liquid chromatography coupled to linear ion trap mass spectrometry for the identification of drug metabolites in biological samples. Rapid Commun. Mass Spectrom. 20, 1351–1360 (2006).
Wagner, C., Sefkow, M. & Kopka, J. Construction and application of a mass spectral and retention time index database generated from plant GC/EI-TOF-MS metabolite profiles. Phytochemistry 62, 887–900 (2003).
Jonsson, P. et al. A strategy for identifying differences in large series of metabolomic samples analyzed by GC/MS. Anal. Chem. 76, 1738–1745 (2004).
Vorst, O. et al. A non-directed approach to the differential analysis of multiple LC–MS-derived metabolic profiles. Metabolomics 1, 169–180 (2005).
Ward, J.L., Harris, C., Lewis, J. & Beale, M.H. Assessment of H-1 NMR spectroscopy and multivariate analysis as a technique for metabolite fingerprinting of Arabidopsis thaliana. Phytochemistry 62, 949–957 (2003).
Allen, J. et al. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat. Biotechnol. 21, 692–696 (2003).
Scholz, M., Gatzek, S., Sterling, A., Fiehn, O. & Selbig, J. Metabolite fingerprinting: detecting biological features by independent component analysis. Bioinformatics 20, 2447–2454 (2004).
Aharoni, A. et al. Nontargeted metabolome analysis by use of Fourier Transform Ion Cyclotron Mass Spectrometry. OMICS 6, 217–234 (2002).
Smedsgaard, J. & Frisvad, J.C. Using direct electrospray mass spectrometry in taxonomy and secondary metabolite profiling of crude fungal extracts. J Microbiol. Methods 25, 5–17 (1996).
Dunn, W.B., Bailey, N.J. & Johnson, H.E. Measuring the metabolome: current analytical technologies. Analyst 130, 606–625 (2005).
Beckmann, M., Parker, D., Enot, D.P., Duval, E. & Draper, J. High-throughput, nontargeted metabolite fingerprinting using nominal mass flow injection electrospray mass spectrometry. Nat. Protoc. 3, 486–504 (2008).
Overy, D.P. et al. Explanatory signal interpretation and metabolite identification strategies for nominal mass FIE-MS metabolite fingerprints. Nat. Protoc. 3, 471–485 (2008).
Parker, D. et al. Rice blast infection of Brachypodium distachyon as a model system to study dynamic host/pathogen interactions. Nat. Protoc. 3, 435–445 (2008).
Enot, D.P., Beckmann, M. & Draper, J. Detecting a difference—assessing generalisability when modelling metabolome fingerprint data in longer term studies of genetically modified plants. Metabolomics 3, 335–347 (2007).
Enot, D.P. & Draper, J. Statistical measures for testing substantial equivalence of GM plant genotypes in a multivariate context. Metabolomics 3, 349–355 (2007).
Jain, A.K., Murty, M.N. & Flynn, P.J. Data clustering: a review. ACM Computing Surveys (CSUR) 31, 264–323 (1999).
Manly, B.F.J. Multivariate Statistical Methods: A Primer. Chapman & Hall/CRC, London (2004).
Zhang, C., Lu, X. & Zhang, X. Significance of gene ranking for classification of microarray samples. EEE/ACM Transactions on Computational Biology and Bioinformatics 3, 312–320 (2006).
Ransohoff, D.F. Rules of evidence for cancer molecular-marker discovery and validation. Nat. Rev. Cancer 4, 309–313 (2004).
Davis, C.A. et al. Reliable gene signatures for microarray classification: assessment of stability and performance. Bioinformatics 22, 2356–2363 (2006).
Wu, B. et al. Comparison of statistical methods for classification of ovarian cancer using mass spectrometry data. Bioinformatics 19, 1636–1643 (2003).
Cristianini, N. & Shawe-Taylor, J. An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods. Cambridge University Press, Cambridge (2000).
Zhu, C., Kitagawa, H. & Faloutsos, C. Example-based outlier detection for high dimensional datasets. IPSJ Digital Courier 1, 234–243 (2005).
Craig, A., Cloarec, O., Holmes, E., Nicholson, J.K. & Lindon, J.C. Scaling and normalization effects in NMR spectroscopic metabonomic data sets. Anal. Chem. 78, 2262–2267 (2006).
Hastie, T., Tibshirani, R. & Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction. Springer Series in Statistics, New York (2001).
Good, P. Permutation Tests: A Practical Guide to Resampling Methods for Testing Hypotheses. Springer Series in Statistics, Heidelberg (2000).
Efron, B. Estimating the error rate of a prediction rule: improvement on cross-validation. J. Am. Stat. Assoc. 78, 316–331 (1983).
Sing, T., Sander, O., Beerenwinkel, N. & Lengauer, T. ROCR: visualizing classifier performance in R. Bioinformatics 21, 3940–3941 (2005).
Fu, W.J., Carroll, R.J. & Wang, S. Estimating misclassification error with small samples via bootstrap cross-validation. Bioinformatics 21, 1979–1986 (2005).
Thomaz, C.E. et al. Using a maximum uncertainty LDA-based approach to classify and analyse MR brain images. Lecture Notes in Computer Science: Medical Image Computing and Computer-Assisted Intervention—MICCAI 2004, pp 291–3 Springer, Berlin, 291–300 (2004).
Yang, J. & Yang, J. Why can LDA be performed in PCA transformed space? Pattern Recognition 36, 563–566 (2003).
Breiman, L. Random Forests. Machine Learning 45, 5–32 (2001).
Zar, J.H. Biostatistics. 2nd edn. (Prentice-Hall, Englewood Cliffs, New Jersey, 1984).
Dietterich, T.G. Ensemble methods in machine learning. Lecture Notes in Computer Science 1857, 1–15 (2000).
Vaidyanathan, S., Kell, D.B. & Goodacre, R. Flow-injection electrospray ionization mass spectrometry of crude cell extracts for high-throughput bacterial identification. J. Am. Soc. Mass Spectrom. 13, 118–128 (2002).
Roessner, U. & Luedemann, A. et al. Metabolic profiling allows comprehensive phenotyping of genetically or environmentally modified plant systems. Plant Cell 13, 11–29 (2001).
Mazzella, N. et al. Use of electrospray ionization mass spectrometry for profiling of crude oil effects on the phospholipid molecular species of two marine bacteria. Rapid Commun. Mass Spectrom. 19, 3579–3588 (2005).
Favretto, D., Piovan, A., Filippini, R. & Caniato, R. Monitoring the production yields of vincristine and vinblastine in Catharanthus roseus from somatic embryogenesis. Semiquantitative determination by flow-injection electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 15, 364–369 (2001).
Rashed, M.S., Al-Ahaidib, L.Y., Aboul-Enein, H.Y., Al-Amoudi, M. & Jacob, M. Determination of L-pipecolic acid in plasma using chiral liquid chromatography-electrospray tandem mass spectrometry. Clin. Chem. 47, 2124–2130 (2001).
Overy, S.A. et al. Application of metabolite profiling to the identification of traits in a population of tomato introgression lines. J. Exp. Bot. 56, 287–296 (2005).
Goodacre, R., York, E.V., Heald, J.K. & Scott, I.M. Chemometric discrimination of unfractionated plant extracts analyzed by electrospray mass spectrometry. Phytochemistry 62, 859–863 (2003).
Koulman, A. et al. High-throughput direct-infusion ion trap mass spectrometry: a new method for metabolomics. Rapid Commun. Mass Spectrom. 21, 421–428 (2007).
Martinez, A.M. & Kak, A.C. PCA versus LDA. IEEE Transactions on: Pattern Analysis and Machine Intelligence 23, 228–233 (2001).
Windeatt, T. Vote counting measures for ensemble classifiers. Pattern Recognition 36, 2743–2756 (2003).
R_Development_Core_Team. R. A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria, ISBN 3-900051-900007-900050, URL http://www.R-project.org (2006).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 57, 289–300 (1995).
Storey, J.D. A direct approach to false discovery rates. J. R. Stat. Soc. Ser. B Stat. Methodol. 64, 479–498 (2002).
Acknowledgements
Financial support was provided for W.L., M.B. and D.P.O by the UK Food Standards Agency G03012 programme. D.P.E. and D.P. were funded by grants MET20483 and BBD0069531 respectively, from the UK Biotechnology and Biological Sciences Research Council.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Enot, D., Lin, W., Beckmann, M. et al. Preprocessing, classification modeling and feature selection using flow injection electrospray mass spectrometry metabolite fingerprint data. Nat Protoc 3, 446–470 (2008). https://doi.org/10.1038/nprot.2007.511
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2007.511
This article is cited by
-
Developing a machine learning model for accurate nucleoside hydrogels prediction based on descriptors
Nature Communications (2024)
-
Spectral binning as an approach to post-acquisition processing of high resolution FIE-MS metabolome fingerprinting data
Metabolomics (2022)
-
Plasma metabolomic profiling in patients with rheumatoid arthritis identifies biochemical features predictive of quantitative disease activity
Arthritis Research & Therapy (2021)
-
Specificity of metabolic colorectal cancer biomarkers in serum through effect size
Metabolomics (2020)
-
Addressing the pitfalls when designing intervention studies to discover and validate biomarkers of habitual dietary intake
Metabolomics (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.