Abstract
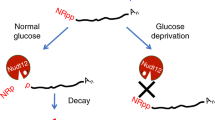
RNA capping and decapping are thought to be distinctive features of eukaryotes. The redox cofactor NAD was recently discovered to be attached to small regulatory RNAs in bacteria in a cap-like manner, and Nudix hydrolase NudC was found to act as a NAD-decapping enzyme in vitro and in vivo. Here, crystal structures of Escherichia coli NudC in complex with substrate NAD and with cleavage product NMN reveal the catalytic residues lining the binding pocket and principles underlying molecular recognition of substrate and product. Biochemical mutation analysis identifies the conserved Nudix motif as the catalytic center of the enzyme, which needs to be homodimeric, as the catalytic pocket is composed of amino acids from both monomers. NudC is single-strand specific and has a purine preference for the 5′-terminal nucleotide. The enzyme strongly prefers NAD-linked RNA (NAD–RNA) over NAD and binds to a diverse set of cellular RNAs in an unspecific manner.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Topisirovic, I., Svitkin, Y.V., Sonenberg, N. & Shatkin, A.J. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip. Rev. RNA 2, 277–298 (2011).
Arribas-Layton, M., Wu, D., Lykke-Andersen, J. & Song, H. Structural and functional control of the eukaryotic mRNA decapping machinery. Biochim. Biophys. Acta 1829, 580–589 (2013).
Chen, Y.G., Kowtoniuk, W.E., Agarwal, I., Shen, Y. & Liu, D.R. LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat. Chem. Biol. 5, 879–881 (2009).
Cahová, H., Winz, M.L., Höfer, K., Nübel, G. & Jäschke, A. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature 519, 374–377 (2015).
Jäschke, A., Höfer, K., Nübel, G. & Frindert, J. Cap-like structures in bacterial RNA and epitranscriptomic modification. Curr. Opin. Microbiol. 30, 44–49 (2016).
Luciano, D.J. & Belasco, J.G. NAD in RNA: unconventional headgear. Trends Biochem. Sci. 40, 245–247 (2015).
Marbaniang, C.N. & Vogel, J. Emerging roles of RNA modifications in bacteria. Curr. Opin. Microbiol. 30, 50–57 (2016).
Mackie, G.A. Ribonuclease E is a 5′-end-dependent endonuclease. Nature 395, 720–723 (1998).
Deana, A., Celesnik, H. & Belasco, J.G. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature 451, 355–358 (2008).
McLennan, A.G. The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 63, 123–143 (2006).
McLennan, A.G. Substrate ambiguity among the Nudix hydrolases: biologically significant, evolutionary remnant, or both? Cell. Mol. Life Sci. 70, 373–385 (2013).
Song, M.G., Bail, S. & Kiledjian, M. Multiple Nudix family proteins possess mRNA decapping activity. RNA 19, 390–399 (2013).
She, M. et al. Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe. Nat. Struct. Mol. Biol. 13, 63–70 (2006).
Frick, D.N. & Bessman, M.J. Cloning, purification, and properties of a novel NADH pyrophosphatase. Evidence for a nucleotide pyrophosphatase catalytic domain in MutT-like enzymes. J. Biol. Chem. 270, 1529–1534 (1995).
Aglietti, R.A., Floor, S.N., McClendon, C.L., Jacobson, M.P. & Gross, J.D. Active site conformational dynamics are coupled to catalysis in the mRNA decapping enzyme Dcp2. Structure 21, 1571–1580 (2013).
Olejnik, K. et al. Mutational analysis of the AtNUDT7 Nudix hydrolase from Arabidopsis thaliana reveals residues required for protein quaternary structure formation and activity. Acta Biochim. Pol. 56, 291–300 (2009).
O'Handley, S.F., Frick, D.N., Dunn, C.A. & Bessman, M.J. Orf186 represents a new member of the Nudix hydrolases, active on adenosine(5′)triphospho(5′)adenosine, ADP-ribose, and NADH. J. Biol. Chem. 273, 3192–3197 (1998).
Deshmukh, M.V. et al. mRNA decapping is promoted by an RNA-binding channel in Dcp2. Mol. Cell 29, 324–336 (2008).
She, M. et al. Structural basis of dcp2 recognition and activation by dcp1. Mol. Cell 29, 337–349 (2008).
Vasilyev, N. & Serganov, A. Structures of RNA complexes with the Escherichia coli RNA pyrophosphohydrolase RppH unveil the basis for specific 5′-end-dependent mRNA decay. J. Biol. Chem. 290, 9487–9499 (2015).
Bessman, M.J., Frick, D.N. & O'Handley, S.F. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 271, 25059–25062 (1996).
Lee, C.R., Kim, M., Park, Y.H., Kim, Y.R. & Seok, Y.J. RppH-dependent pyrophosphohydrolysis of mRNAs is regulated by direct interaction with DapF in Escherichia coli. Nucleic Acids Res. 42, 12746–12757 (2014).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Laskowski, R.A., Macarthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Halbritter, F., Vaidya, H.J. & Tomlinson, S.R. GeneProf: analysis of high-throughput sequencing experiments. Nat. Methods 9, 7–8 (2012).
Kersey, P.J. et al. Ensembl Genomes 2016: more genomes, more complexity. Nucleic Acids Res. 44, D1, D574–D580 (2016).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Höfer, K., Abele, F., Schlotthauer, J. & Jäschke, A. Synthesis of 5′-NAD-capped RNA. Bioconjug. Chem. 27, 874–877 (2016).
Acknowledgements
We are grateful to L. Obenauer (Heidelberg University) for assistance with NGS data analysis, L. Kiss for experimental assistance and to W. Shi at beamline X29A at the Brookhaven National Laboratory for support in diffraction data collection. A.J. is supported by the German Research Foundation (DFG SPP 1784, grant JA 794/10-1) and by the Baden-Württemberg Stiftung (BWST-NCRNA-045). The structural research was supported by funds from US National Institutes of Health (NIH) grant GM104962 to D.J.P. and the Thousand Young Talent Program of China and the Chinese Academy of Sciences to J.D.
Author information
Authors and Affiliations
Contributions
K.H., S.L., D.J.P., and A.J. designed research; S.L. and J.D. performed the crystallographic investigation; K.H., F.A., J.F., J.S., and J.G. carried out the functional studies on NudC; all authors analyzed data; K.H., S.L., J.F., J.D., D.J.P., and A.J. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–8 and Supplementary Tables 1 and 2. (PDF 3030 kb)
Rights and permissions
About this article
Cite this article
Höfer, K., Li, S., Abele, F. et al. Structure and function of the bacterial decapping enzyme NudC. Nat Chem Biol 12, 730–734 (2016). https://doi.org/10.1038/nchembio.2132
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2132
This article is cited by
-
Identification of NAD-RNA species and ADPR-RNA decapping in Archaea
Nature Communications (2023)
-
Xrn1 is a deNADding enzyme modulating mitochondrial NAD-capped RNA
Nature Communications (2022)
-
Extensive 5′-surveillance guards against non-canonical NAD-caps of nuclear mRNAs in yeast
Nature Communications (2020)
-
Dinucleoside polyphosphates act as 5′-RNA caps in bacteria
Nature Communications (2020)
-
The first comprehensive phylogenetic and biochemical analysis of NADH diphosphatases reveals that the enzyme from Tuber melanosporum is highly active towards NAD+
Scientific Reports (2019)