Abstract
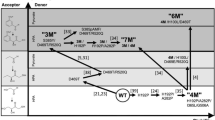
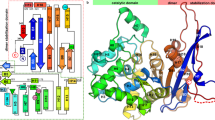
The protein databases contain many proteins with unknown function. A computational approach for predicting ligand specificity that requires only the sequence of the unknown protein would be valuable for directing experiment-based assignment of function. We focused on a family of unknown proteins in the mechanistically diverse enolase superfamily and used two approaches to assign function: (i) enzymatic assays using libraries of potential substrates, and (ii) in silico docking of the same libraries using a homology model based on the most similar (35% sequence identity) characterized protein. The results matched closely; an experimentally determined structure confirmed the predicted structure of the substrate-liganded complex. We assigned the N-succinyl arginine/lysine racemase function to the family, correcting the annotation (L-Ala-D/L-Glu epimerase) based on the function of the most similar characterized homolog. These studies establish that ligand docking to a homology model can facilitate functional assignment of unknown proteins by restricting the identities of the possible substrates that must be experimentally tested.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Friedberg, I., Jambon, M. & Godzik, A. New avenues in protein function prediction. Protein Sci. 15, 1527–1529 (2006).
Watson, J.D. et al. Towards fully automated structure-based function prediction in structural genomics: a case study. J. Mol. Biol. 367, 1511–1522 (2007).
Adams, M., Joachimiak, A., Kim, R., Montelione, G.T. & Norvell, J. Meeting review: 2003 NIH protein structure initiative workshop in protein production and crystallization for structural and functional genomics. J. Struct. Funct. Genomics 5, 1–2 (2004).
Lattman, E. The state of the Protein Structure Initiative. Proteins 54, 611–615 (2004).
Norvell, J.C. & Machalek, A.Z. Structural genomics programs at the US National Institute of General Medical Sciences. Nat. Struct. Biol. 7 (suppl.), 931 (2000).
Babbitt, P.C. et al. The enolase superfamily: a general strategy for enzyme-catalyzed abstraction of the α-protons of carboxylic acids. Biochemistry 35, 16489–16501 (1996).
Gerlt, J.A., Babbitt, P.C. & Rayment, I. Divergent evolution in the enolase superfamily: the interplay of mechanism and specificity. Arch. Biochem. Biophys. 433, 59–70 (2005).
Pegg, S.C. et al. Leveraging enzyme structure-function relationships for functional inference and experimental design: the structure-function linkage database. Biochemistry 45, 2545–2555 (2006).
Hasson, M.S. et al. Evolution of an enzyme active site: the structure of a new crystal form of muconate lactonizing enzyme compared with mandelate racemase and enolase. Proc. Natl. Acad. Sci. USA 95, 10396–10401 (1998).
Thompson, T.B. et al. Evolution of enzymatic activity in the enolase superfamily: structure of o-succinylbenzoate synthase from Escherichia coli in complex with Mg(II) and o-succinylbenzoate. Biochemistry 39, 10662–10676 (2000).
Schmidt, D.M., Hubbard, B.K. & Gerlt, J.A. Evolution of enzymatic activities in the enolase superfamily: functional assignment of unknown proteins in Bacillus subtilis and Escherichia coli as L-Ala-D/L-Glu epimerases. Biochemistry 40, 15707–15715 (2001).
Gulick, A.M., Schmidt, D.M., Gerlt, J.A. & Rayment, I. Evolution of enzymatic activities in the enolase superfamily: crystal structures of the L-Ala-D/L-Glu epimerases from Escherichia coli and Bacillus subtilis. Biochemistry 40, 15716–15724 (2001).
Taylor Ringia, E.A. et al. Evolution of enzymatic activity in the enolase superfamily: functional studies of the promiscuous o-succinylbenzoate synthase from Amycolatopsis. Biochemistry 43, 224–229 (2004).
Thoden, J.B. et al. Evolution of enzymatic activity in the enolase superfamily: structural studies of the promiscuous o-succinylbenzoate synthase from Amycolatopsis. Biochemistry 43, 5716–5727 (2004).
Sakai, A. et al. Evolution of enzymatic activities in the enolase superfamily: N-succinylamino acid racemase and a new pathway for the irreversible conversion of D- to L-amino acids. Biochemistry 45, 4455–4462 (2006).
Glasner, M.E. et al. Evolution of structure and function in the o-succinylbenzoate synthase/N-acylamino acid racemase family of the enolase superfamily. J. Mol. Biol. 360, 228–250 (2006).
Palmer, D.R. et al. Unexpected divergence of enzyme function and sequence: “N-acylamino acid racemase” is o-succinylbenzoate synthase. Biochemistry 38, 4252–4258 (1999).
Klenchin, V.A., Schmidt, D.M., Gerlt, J.A. & Rayment, I. Evolution of enzymatic activities in the enolase superfamily: structure of a substrate-liganded complex of the L-Ala-D/L-Glu epimerase from Bacillus subtilis. Biochemistry 43, 10370–10378 (2004).
Kalyanaraman, C., Bernacki, K. & Jacobson, M.P. Virtual screening against highly charged active sites: identifying substrates of alpha-beta barrel enzymes. Biochemistry 44, 2059–2071 (2005).
Sherman, W., Day, T., Jacobson, M.P., Friesner, R.A. & Farid, R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 49, 534–553 (2006).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Jacobson, M.P., Friesner, R.A., Xiang, Z. & Honig, B. On the role of the crystal environment in determining protein side-chain conformations. J. Mol. Biol. 320, 597–608 (2002).
Otwinowski, Z. & Minor, W. in Methods in Enzymology Vol. 276 (eds. Carter, C.W.J., Sweet, R.M., Abelson, J.N. & Simon, M.I.) 307–326 (Academic, New York, 1997).
McCoy, A.J., Grosse-Kunstleve, R.W., Storoni, L.C. & Read, R.J. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 (2005).
Perrakis, A., Morris, R. & Lamzin, V.S. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6, 458–463 (1999).
Jones, T.A. Diffraction methods for biological macromolecules. Interactive computer graphics: FRODO. Methods Enzymol. 115, 157–171 (1985).
Brunger, A.T. et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Schuttelkopf, A.W. & van Aalten, D.M. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 (2004).
Acknowledgements
This work was supported by the US National Institutes of Health (1 P01 GM-71790).
Author information
Authors and Affiliations
Contributions
L.S. isolated BC0371 and performed library screening and kinetic assays; C.K. performed molecular modeling and in silico library docking; A.A.F. and E.V.F. crystallized BC0371 and solved its structure; M.E.G. and S.B. performed sequence alignments and generated the Cytoscape-based representation of sequence relationships; H.J.I. cloned the gene encoding BC0371 and spectroscopically characterized dipeptide substrates; P.C.B., S.C.A., M.P.J. and J.A.G. designed the experiments and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
M.P.J. is a member of the Scientific Advisory Board of Schrodinger Inc.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–3, Supplementary Table 1 and Supplementary Methods (PDF 1201 kb)
Rights and permissions
About this article
Cite this article
Song, L., Kalyanaraman, C., Fedorov, A. et al. Prediction and assignment of function for a divergent N-succinyl amino acid racemase. Nat Chem Biol 3, 486–491 (2007). https://doi.org/10.1038/nchembio.2007.11
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2007.11
This article is cited by
-
Biochemical and Mutational Characterization of N-Succinyl-Amino Acid Racemase from Geobacillus stearothermophilus CECT49
Molecular Biotechnology (2015)
-
Computational evaluation of factors governing catalytic 2-keto acid decarboxylation
Journal of Molecular Modeling (2014)
-
Discovery of new enzymes and metabolic pathways by using structure and genome context
Nature (2013)
-
Dynamic changes in the secondary structure of ECE-1 and XCE account for their different substrate specificities
BMC Bioinformatics (2012)
-
A decade of chemical biology
Nature Chemical Biology (2010)