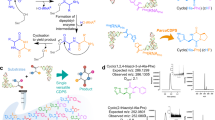
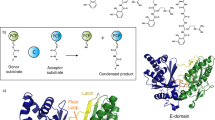
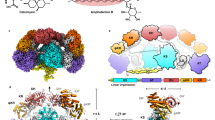
Abstract
Cyclodipeptide synthases (CDPSs) constitute a family of peptide bond–forming enzymes that use aminoacyl-tRNAs for the synthesis of cyclodipeptides. Here, we describe the activity of 41 new CDPSs. We also show that CDPSs can be classified into two main phylogenetically distinct subfamilies characterized by specific functional subsequence signatures, named NYH and XYP. All 11 previously characterized CDPSs belong to the NYH subfamily, suggesting that further special features may be yet to be discovered in the other subfamily. CDPSs synthesize a large diversity of cyclodipeptides made up of 17 proteinogenic amino acids. The identification of several CDPSs having the same specificity led us to determine specificity sequence motifs that, in combination with the phylogenetic distribution of CDPSs, provide a first step toward being able to predict the cyclodipeptides synthesized by newly discovered CDPSs. The determination of the activity of ten more CDPSs with predicted functions constitutes a first experimental validation of this predictive approach.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Gondry, M. et al. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes. Nat. Chem. Biol. 5, 414–420 (2009).
Vetting, M.W., Hegde, S.S. & Blanchard, J.S. The structure and mechanism of the Mycobacterium tuberculosis cyclodityrosine synthetase. Nat. Chem. Biol. 6, 797–799 (2010).
Sauguet, L. et al. Cyclodipeptide synthases, a family of class-I aminoacyl-tRNA synthetase-like enzymes involved in non-ribosomal peptide synthesis. Nucleic Acids Res. 39, 4475–4489 (2011).
Bonnefond, L. et al. Structural basis for nonribosomal peptide synthesis by an aminoacyl-tRNA synthetase paralog. Proc. Natl. Acad. Sci. USA 108, 3912–3917 (2011).
Seguin, J. et al. Nonribosomal peptide synthesis in animals: the cyclodipeptide synthase of Nematostella. Chem. Biol. 18, 1362–1368 (2011).
Moutiez, M. et al. Unravelling the mechanism of non-ribosomal peptide synthesis by cyclodipeptide synthases. Nat. Commun. 5, 5141 (2014).
Moutiez, M. et al. Specificity determinants for the two tRNA substrates of the cyclodipeptide synthase AlbC from Streptomyces noursei. Nucleic Acids Res. 42, 7247–7258 (2014).
Giessen, T.W., von Tesmar, A.M. & Marahiel, M.A. A tRNA-dependent two-enzyme pathway for the generation of singly and doubly methylated ditryptophan 2,5-diketopiperazines. Biochemistry 52, 4274–4283 (2013).
Giessen, T.W., von Tesmar, A.M. & Marahiel, M.A. Insights into the generation of structural diversity in a tRNA-dependent pathway for highly modified bioactive cyclic dipeptides. Chem. Biol. 20, 828–838 (2013).
Aravind, L., de Souza, R.F. & Iyer, L.M. Predicted class-I aminoacyl tRNA synthetase-like proteins in non-ribosomal peptide synthesis. Biol. Direct 5, 48 (2010).
Belin, P. et al. The nonribosomal synthesis of diketopiperazines in tRNA-dependent cyclodipeptide synthase pathways. Nat. Prod. Rep. 29, 961–979 (2012).
Giessen, T.W. & Marahiel, M.A. The tRNA-dependent biosynthesis of modified cyclic dipeptides. Int. J. Mol. Sci. 15, 14610–14631 (2014).
Stachelhaus, T., Mootz, H.D. & Marahiel, M.A. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6, 493–505 (1999).
Challis, G.L., Ravel, J. & Townsend, C.A. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7, 211–224 (2000).
Tang, M.R., Sternberg, D., Behr, R.K., Sloma, A. & Berka, R.M. Use of transcriptional profiling & bioinformatics to solve production problems. Ind. Biotechnol. (New Rochelle N.Y.) 2, 66–74 (2006).
Fukushima, K., Yazawa, K. & Arai, T. Biological activities of albonoursin. J. Antibiot. (Tokyo) 26, 175–176 (1973).
Lautru, S., Gondry, M., Genet, R. & Pernodet, J.L. The albonoursin gene cluster of S. noursei: biosynthesis of diketopiperazine metabolites independent of nonribosomal peptide synthetases. Chem. Biol. 9, 1355–1364 (2002).
Gu, B., He, S., Yan, X. & Zhang, L. Tentative biosynthetic pathways of some microbial diketopiperazines. Appl. Microbiol. Biotechnol. 97, 8439–8453 (2013).
Gondry, M. et al. Cyclic dipeptide oxidase from Streptomyces noursei. Isolation, purification and partial characterization of a novel amino acyl alpha,beta-dehydrogenase. Eur. J. Biochem. 268, 1712–1721 (2001).
Belin, P. et al. Identification and structural basis of the reaction catalyzed by CYP121, an essential cytochrome P450 in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 106, 7426–7431 (2009).
Cryle, M.J., Bell, S.G. & Schlichting, I. Structural and biochemical characterization of the cytochrome P450 CypX (CYP134A1) from Bacillus subtilis: a cyclo-L-leucyl-L-leucyl dipeptide oxidase. Biochemistry 49, 7282–7296 (2010).
Sambrook, J., Fritsch, E.F. & Maniatis, T. Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, 2001).
Altschul, S.F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Schäffer, A.A. et al. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 29, 2994–3005 (2001).
Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Gouy, M., Guindon, S. & Gascuel, O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27, 221–224 (2010).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23, 127–128 (2007).
Letunic, I. & Bork, P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478 (2011).
Remmert, M., Biegert, A., Hauser, A. & Soding, J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat. Methods 9, 173–175 (2012).
Schneider, T.D. & Stephens, R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 (1990).
Crooks, G.E., Hon, G., Chandonia, J.M. & Brenner, S.E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Studier, F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005).
Braud, S. et al. Dual expression system suitable for high-throughput fluorescence-based screening and production of soluble proteins. J. Proteome Res. 4, 2137–2147 (2005).
Bajad, S.U. et al. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J. Chromatogr. A 1125, 76–88 (2006).
Chen, Y.-H., Liou, S.-E. & Chen, C.-C. Two-step mass spectrometric approach for the identification of diketopiperazines in chicken essence. Eur. Food Res. Technol. 218, 589–597 (2004).
Falick, A.M., Hines, W.M., Medzihradszly, K.F., Baldwin, M.A. & Gibson, B.W. Low-mass ions produced from peptides by high-energy collision-induced dissociation in tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 4, 882–893 (1993).
Johnson, R.S., Martin, S.A., Biemann, K., Stults, J.T. & Watson, J.T. Novel fragmentation process of peptides by collision-induced decomposition in a tandem mass spectrometer: differentiation of leucine and isoleucine. Anal. Chem. 59, 2621–2625 (1987).
Papayannopoulos, I.A. The interpretation of collision-induced dissociation tandem mass spectra of peptides. Mass Spectrom. Rev. 14, 49–73 (1995).
Roepstorff, P. & Fohlman, J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom. 11, 601 (1984).
Stark, T. & Hofmann, T. Structures, sensory activity, and dose/response functions of 2,5-diketopiperazines in roasted cocoa nibs (Theobroma cacao). J. Agric. Food Chem. 53, 7222–7231 (2005).
Armirotti, A., Millo, E. & Damonte, G. How to discriminate between leucine and isoleucine by low energy ESI-TRAP MSn. J. Am. Soc. Mass Spectrom. 18, 57–63 (2007).
Jeedigunta, S., Krenisky, J.M. & Kerr, R.G. Diketopiperazines as advanced intermediates in the biosynthesis of Ecteinascidins. Tetrahedron 56, 3303–3307 (2000).
Acknowledgements
This work was supported by the CEA, the CNRS, the Paris-Sud University and a grant from the French National Research Agency (ANR 2010/Blan 1501 01) to M.G. and J.-L.P. I.B.J. was supported by a doctoral fellowship from the CEA. The Service d'Ingénierie Moléculaire des Protéines is member of the Laboratory of Excellence LERMIT. We warmly thank V. Dive, head of the Service d'Ingénierie Moléculaire des Protéines, for his continuous support and encouragement throughout this work. We thank O. Lespinet for advice about the building of phylogenetic trees, D. Vallenet, M. Stam and M. Sorokina for helpful discussion on bioinformatics, and A. Ponties for technical assistance in cloning experiments. We are indebted to L. Beuvier for skillful assistance in mass data analysis. We thank F. Fenaille for kindly performing the experiments using the Orbitrap mass spectrophotometer. We thank P. Kessler and O. Lequin for skillful assistance in NMR experiments.
Author information
Authors and Affiliations
Contributions
M.G. obtained funding. M.M., J.-L.P., M.G. and P.B. developed the hypothesis and designed the study. I.B.J., M.M. and P.B. performed bioinformatic analyses. I.B.J., J.W., E.D. and C.M. performed cloning experiments and prepared culture supernatants. I.B.J. performed LC/MS/MS analysis. I.B.J. and M.M. analyzed MS/MS data. I.B.J., J.S., E.F. and P.B. purified cyclodipeptides from culture supernatants. S.D. performed amino acid composition analyses. A.L. and S.D. chemically synthesized cyclodipeptides. R.T. performed high-resolution mass spectrometry. I.B.J., M.M., J.W., E.D., J.S., J.L.P., M.G. and P.B. analyzed and discussed the results. I.B.J., M.M., M.G. and P.B. prepared the draft manuscript. All of the authors participated in the production of the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–3 and Supplementary Figures 1–7. (PDF 1651 kb)
Supplementary Data Set 1
Database information and sequence data relative to each putative CDPS produced in this study. (XLSX 46 kb)
Supplementary Data Set 2
Cyclodipeptides detected in bacterial culture supernatants. (XLSX 57 kb)
Supplementary Data Set 3
Sequence identity between the 11 previously identified CDPSs and the 49 CDPSs produced in this study. Sequence identities were obtained from the percent identity matrix created with Clustal Omega. (XLSX 64 kb)
Supplementary Data Set 4
HHPred results for the selected CDPSs. (XLSX 38 kb)
Supplementary Data Set 5
Multiple sequence alignment of the 49 selected CDPSs plus the 11 biochemically characterized CDPSs (FASTA format). Inactive CDPSs are identified by an asterisk. (PDF 501 kb)
Supplementary Data Set 6
N1-N72 bases of available tRNAs of the organisms containing the CDPSs studied. (XLSX 30 kb)
Supplementary Data Set 7
Prediction of aminoacyl-binding pockets in CDPSs. (XLSX 20 kb)
Supplementary Data Set 8
Phylogenetic tree of CDPS sequences retrieved by database searches (September 2014). (PDF 3726 kb)
Supplementary Data Set 9
CDPS groups according to clustering on the phylogenetic tree and identity between the P1 and P2 binding pockets. (XLSX 343 kb)
Rights and permissions
About this article
Cite this article
Jacques, I., Moutiez, M., Witwinowski, J. et al. Analysis of 51 cyclodipeptide synthases reveals the basis for substrate specificity. Nat Chem Biol 11, 721–727 (2015). https://doi.org/10.1038/nchembio.1868
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1868