Abstract
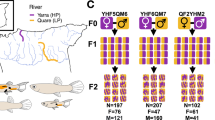
Relative to morphological traits, we know little about how genetics influence the evolution of complex behavioural differences in nature1. It is unclear how the environment influences natural variation in heritable behaviour2, and whether complex behavioural differences evolve through few genetic changes, each affecting many aspects of behaviour, or through the accumulation of several genetic changes that, when combined, give rise to behavioural complexity3. Here we show that in nature, oldfield mice (Peromyscus polionotus) build complex burrows with long entrance and escape tunnels, and that burrow length is consistent across populations, although burrow depth varies with soil composition. This burrow architecture is in contrast with the small, simple burrows of its sister species, deer mice (P. maniculatus). When investigated under laboratory conditions, both species recapitulate their natural burrowing behaviour. Genetic crosses between the two species reveal that the derived burrows of oldfield mice are dominant and evolved through the addition of multiple genetic changes. In burrows built by first-generation backcross mice, entrance-tunnel length and the presence of an escape tunnel can be uncoupled, suggesting that these traits are modular. Quantitative trait locus analysis also indicates that tunnel length segregates as a complex trait, affected by at least three independent genetic regions, whereas the presence of an escape tunnel is associated with only a single locus. Together, these results suggest that complex behaviours—in this case, a classic ‘extended phenotype’4—can evolve through multiple genetic changes each affecting distinct behaviour modules.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Boake, C. R. B. et al. Genetic tools for studying adaptation and the evolution of behavior. Am. Nat. 160, S143–S159 (2002)
West-Eberhard, M. J. Developmental Plasticity and Evolution (Oxford Univ. Press, 2003)
Mackay, T. F. C. The genetic architecture of quantitative traits. Annu. Rev. Genet. 35, 303–339 (2001)
Dawkins, R. The Extended Phenotype (W. H. Freeman, 1982)
Hansell, M. H. Animal Architecture (Oxford Univ. Press, 2005)
Lorenz, K. Z. The evolution of behaviour. Sci. Am. 199, 67–78 (1958)
Peichel, C. L. et al. The genetic architecture of divergence between threespine stickleback species. Nature 414, 901–905 (2001)
Steiner, C. C., Weber, J. N. & Hoekstra, H. E. Adaptive variation in beach mice caused by two interacting pigmentation genes. PLoS Biol. 5, 1880–1889 (2007)
Sumner, F. B. & Karol, J. J. Notes on the burrowing habits of Peromyscus polionotus. J. Mamm. 10, 213–215 (1929)
Hayne, D. W. Burrowing habits of Peromyscus polionotus. J. Mamm. 17, 420–421 (1936)
Rand, A. L. & Host, P. Mammal notes from Highland County, Florida. Bull. Am. Mus. Nat. Hist. 80, 1–21 (1942)
Houtcooper, W. C. Rodent seed supply and burrows of Peromyscus in cultivated fields. Proc. Indiana Acad. Sci. 81, 348–389 (1971)
Schwartz, C. W. & Schwartz, E. R. The Wild Mammals of Missouri (Univ. Missouri Press, 1981)
Baker, R. H. in Biology of Peromyscus (Rodentia) (ed. King, J. A. ) (American Society of Mammalogists, 1968)
Wolfe, J. L. & Esher, R. J. Burrowing behaviour of old-field mice (Peromyscus polionotus): a laboratory investigation. Bio. Behav. 2, 343–351 (1977)
Dawson, W. D., Lake, C. E. & Schumpert, S. S. Inheritance of burrow building in Peromyscus. Behav. Genet. 18, 371–382 (1988)
Weber, J. N. & Hoekstra, H. E. The evolution of burrowing behavior in deer mice. Anim. Behav. 77, 603–609 (2009)
Tan, K. H. Soil Sampling, Preparation, and Analysis (CRC Press, 2005)
Wright, S. A mutation of the guinea pig, tending to restore the pentadactyl foot when heterozygous, producing a monstrosity when homozygous. Genetics 20, 84–107 (1935)
Peterson, B. K., Weber, J. N., Kay, E. H., Fisher, H. S. & Hoekstra, H. E. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7, e37135 (2012)
Beavis, W. D. in Molecular Dissection of Complex Traits (ed. Paterson, A. H. ) 431–528 (CRC Press, 1998)
Bendesky, A. & Bargmann, C. I. Genetic contributions to behaviour at the gene–environment interface. Nature Rev. Genet. 12, 809–820 (2011)
Fitzpatrick, M. J. et al. Candidate genes for behavioural ecology. Trends Ecol. Evol. 20, 96–104 (2005)
Huxley, J. S. The courtship habits of the great crested grebe (Podiceps cristatus) with an addition to the theory of sexual selection. Proc. Zool. Soc. Lond. 35, 253–291 (1914)
Mallarino, R. et al. Two developmental modules establish 3D beak shape variation in Darwin’s finches. Proc. Natl Acad. Sci. USA 108, 4057–4062 (2011)
Xu, X. et al. Modular genetic control of sexually dimorphic behaviors. Cell 148, 596–607 (2012)
Felthauser, M. & McInroy, D. Mapping pocket gopher burrow systems with expanding polyurethane foam. J. Wildl. Manage. 47, 555–558 (1983)
R Development Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2011)
Broman, K. W., Wu, H., Sen, Ś. & Churchill, G. A. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890 (2003)
Lincoln, S. E. & Lander, E. S. Systematic detection of errors in genetic linkage data. Genomics 14, 604–610 (1992)
Doerge, R. W. & Rebai, A. Significance thresholds for QTL interval mapping tests. Heredity 76, 459–464 (1996)
Sen, Ś., Satagopan, J., Broman, K. W. & Churchill, G. A. R/qtlDesign: Inbred Line Cross Experimental Design (UC San Francisco: Center for Bioinformatics and Molecular Biostatistics, 2006)
Cheng, R. et al. Genome-wide association studies and the problem of relatedness among advanced intercross lines and other highly recombinant populations. Genetics 185, 1033–1044 (2010)
Cheng, R., Abney, M., Palmer, A. A. & Skol, A. D. QTLRel: an R package for genome-wide association studies in which relatedness is a concern. BMC Genet. 12, 66 (2011)
Acknowledgements
We thank D. Brimmer, A. Chiu, A. Goldberg, J. Hopwood, W. Tong, S. Wolff and the Hoekstra laboratory for assistance with behavioural assays and animal husbandry; D. Haig, B. Ölveczky, N. E. Pierce and J. Sanes for helpful discussions; and Harvard’s Office of Animal Resources, particularly J. Rocca and M. O’Donnell. We also thank R. Barrett, A. Bendesky, H. Fisher, E. Kay, H. Metz and W. Tong for comments on the manuscript. This research was funded by Chapman Funds for Vertebrate Locomotion to J.N.W., National Science Foundation grant (IOS-0910164) to J.N.W. and H.E.H., and an Arnold and Mabel Beckman Young Investigator Award to H.E.H.
Author information
Authors and Affiliations
Contributions
J.N.W. and H.E.H. conceived and designed the experiments. B.K.P. and J.N.W. generated the ddRAD genotypes. J.N.W. performed the behaviour experiments and analysed the genetic and behavioural data. J.N.W. and H.E.H. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-2 and Supplementary Figures 1-4. (PDF 342 kb)
P. polionotus exiting through an escape tunnel and burrow casting method
P. polionotus erupt from an escape tunnel when intruders enter their burrow. We can take advantage of this behavior in the laboratory to remove mice from their burrows, while keeping the burrow architecture intact. Once empty, burrows size and shape can be quantified by constructing and measuring polyurethane casts of each burrow. (MOV 11455 kb)
Rights and permissions
About this article
Cite this article
Weber, J., Peterson, B. & Hoekstra, H. Discrete genetic modules are responsible for complex burrow evolution in Peromyscus mice. Nature 493, 402–405 (2013). https://doi.org/10.1038/nature11816
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11816
This article is cited by
-
Genetic Analysis of the Stereotypic Phenotype in Peromyscus maniculatus (deer mice)
Behavior Genetics (2023)
-
Morphological evolution and the behavioral organization of agricultural division of labor in the leafcutter ant Atta cephalotes
Behavioral Ecology and Sociobiology (2023)
-
Rethinking the Architecture of Attachment: New Insights into the Role for Oxytocin Signaling
Affective Science (2022)
-
Analysis of the Form-Function Relationship: Digging Behavior as a Case Study
Journal of Mammalian Evolution (2021)
-
Biogeomorphological eco-evolutionary feedback between life and geomorphology: a theoretical framework using fossorial mammals
The Science of Nature (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.