Abstract
Genetic structuring in response to the glacial cycles has been investigated for many plant species, but exclusively high-arctic ones have not been studied. Such extremely cold-adapted species have probably experienced range reductions under the present climate. Here we compare three predominantly selfing species of Draba with different distributions and hardiness (D. subcapitata, high-arctic; D. nivalis, arctic to arctic–alpine; D. fladnizensis, arctic–alpine) for genetic structuring on the basis of two different types of molecular markers (10 microsatellite loci and 160 amplified fragment length polymorphisms (AFLPs)). The degree of genetic structuring within these species is of particular interest because it has been shown that they contain many cryptic biological species. The high-arctic D. subcapitata had less phylogeographic structure, less diversity and fewer private alleles than the other two species, suggesting that long-distance dispersal may occur more frequently in the high arctic, that hardy plants may have higher probability for establishment after dispersal under high-arctic conditions and that high-arctic species may have experienced a bottleneck during the present interglacial. In contrast, D. fladnizensis and D. nivalis showed distinct phylogeographic structure and more diversity, suggesting separate long-term refugia in Eurasia and North America/Beringia. The AFLP markers revealed more phylogeographic structuring than the microsatellites, possibly because of the higher number of loci surveyed and/or because structure at very large geographic scales is blurred by high mutation rate leading to homoplasy at microsatellite loci. The number of genetic groups detected was in any case insignificant compared with the numerous cryptic biological species known within these species, supporting rapid development of sterility barriers.
Similar content being viewed by others
Introduction
The effect of the Pleistocene glaciations on the genetic structure of circumpolar plants has been investigated for several species, for example, Cassiope tetragona, Vaccinium uliginosum, Saxifraga oppositifolia and Rubus chamaemorus (Abbott et al., 2000; Abbott and Comes, 2004; Alsos et al., 2005; Eidesen et al., 2007a, 2007b; Ehrich et al., 2008). Considerable geographic structuring of the genetic diversity has been identified in all of them. Distinct phylogeographic structuring has also been observed in many arctic animals, for example, Lemmus spp., Daphnia pulex and Calidris alpina (Weider et al., 1999; Wennerberg, 2001; Federov et al., 2003). However, in some arctic animals, such as the polar fox (Dalén et al., 2005), the polar bear (Paetkau et al., 1999) and the snowy owl (Marthinsen et al., 2008), there is surprisingly little geographic structuring of the genetic diversity even at the vast circumpolar scale. Thus, it is possible that hardy arctic species can be connected through high levels of gene flow across the entire polar basin, preventing geographic differentiation. It is also most likely that the ranges of the most cold-adapted species have become more restricted under the present climatic conditions than the ranges of less hardy ones, and that they occupied larger areas during colder periods (Stewart and Dalén, 2008). The low levels of genetic diversity observed in the polar fox could, for example, indicate postglacial bottlenecking (Dalén et al., 2005). However, there are no data available for exclusively high-arctic plants. Although plants traditionally are considered less mobile than animals, in particular those able to migrate across frozen sea ice, it has recently been shown that arctic plants can have considerable long-distance dispersal ability (Alsos et al., 2007).
Draba is one of the largest genera in the Brassicaceae, consisting of about 350 species (Al-Shehbaz, 1987). It is a polyploid-rich genus, usually with a chromosome base number of x=8 (Al-Shehbaz, 1987). The plants are small flowered and have seeds without any morphological adaptations for long-distance dispersal. The arctic–alpine D. nivalis group (sensu Mulligan, 1974) contains several diploids and polyploids that probably evolved during the Pleistocene (Grundt et al., 2004). Three of the species, D. fladnizensis Wulfen, D. nivalis Liljeblad and D. subcapitata Simmons, formed distinct lineages in a phylogeny inferred from RAPDs and ITS, but their relationships were not fully resolved (Grundt et al., 2004). These species are predominantly autogamous, exclusively or mainly diploid and have circumpolar distributions (Grundt, 2003). They vary, however, from the exclusively high-arctic and very hardy D. subcapitata through the more broadly arctic to arctic–alpine D. nivalis to the less hardy, arctic–alpine D. fladnizensis, which occurs throughout the arctic as well as in temperate mountains (Figure 1). They are typically restricted to similar, heavily wind- and frost-stressed but quite stable habitats with little competition, such as exposed cliffs, ridges and dry heaths with discontinuous vegetation. The hardiest of them, D. subcapitata, extends way into the barren habitats in the far north.
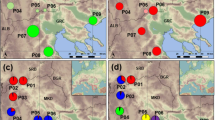
Geographic origin of the 76 Draba populations analyzed and their grouping within each of the three species according to the Bayesian clustering analyses (STRUCTURE) based on nine (D. nivalis and D. subcapitata) or ten (D. fladnizensis) microsatellite loci and 160 AFLP markers. A few populations have been slightly moved to improve visualization. A principal coordinate analysis (PCO) of all individuals, colored according to STRUCTURE group, is shown with each map. The geographic distribution of the species is modified from Hultén and Fries (1986).
Surprisingly, although these three diploids are young and appear ecologically and morphologically uniform throughout their ranges, each of them has recently been found to comprise numerous cryptic biological species (Brochmann et al., 1993; Grundt et al., 2006). The first generation offspring are mostly sterile when crossing individuals from different populations or geographic areas. In the arctic–alpine D. fladnizensis, F1 sterility was significantly correlated with geographic distance between the parents, but in the more arctic D. nivalis, no such correlation was found (Grundt et al., 2006). Weak, but significant, correlation between genetic distance between the parents (on the basis of amplified fragment length polymorphisms (AFLP)) and F1 sterility was found for both D. fladnizensis and D. nivalis (too few crosses were made in the high-arctic D. subcapitata to test for correlations). However, only few AFLP loci were surveyed in this study.
No ideal molecular marker has yet been developed for phylogeographic inference in plants. AFLP markers have frequently been used in plant phylogeography. They may potentially cover large areas of the genome and are easy to use, as no initial DNA sequence information is needed. However, AFLPs are dominant and anonymous, and comigrating fragments are not necessarily homologous. AFLP analysis also depends on high-quality DNA and thus requires fresh material or material immediately dried in silica gel, which is difficult to obtain for species having large and partly inaccessible geographic ranges. In contrast, microsatellites are more expensive to develop but they are codominant, and DNA of sufficient quality can be extracted from herbarium material, making it easier to obtain representative samples from large areas. More work is needed to assess the usefulness of microsatellite markers in phylogeography, especially on larger geographic scales. So far, only a few, mostly regional studies have included direct comparison between AFLP and microsatellite variation. The results suggest that AFLPs may reveal more structure in large data sets than microsatellites (Gaudeul et al., 2004; Woodhead et al., 2005; Sønstebø et al., 2007).
Chloroplast DNA data have also been successfully used for large-scale phylogeographic inference in plants, including arctic ones (for example, Alsos et al., 2005). However, in a pilot study of the Draba nivalis group, sequencing of several chloroplast regions revealed very little variation (Skrede, 2008). In this study, we therefore compare genetic structuring and diversity in the three circumpolar species of Draba (D. fladnizensis, D. nivalis and D. subcapitata) in their entire range on the basis of AFLPs and microsatellites. First, we ask whether levels of geographic structuring and diversity are associated with hardiness and abundance in the northernmost areas. We expect that the most hardy, high-arctic species (D. subcapitata) may show less structuring than the other two species because its higher population densities at the highest latitudes may provide more seeds available for long-distance dispersal across the high-arctic open landscapes, and because its hardiness increases the probability for successful establishment after dispersal (cf. Alsos et al., 2007). We also expect that this very hardy species shows less diversity because of more heavy bottlenecking associated with postglacial warming. Second, we ask whether the two types of molecular markers, AFLPs and microsatellites (recently developed for Draba by Skrede et al., 2008b), provide similar information on degree of genetic structuring and diversity in these three species. Third, by extending the number of loci surveyed compared with Grundt et al. (2006; by including microsatellite data and increasing the number of AFLP loci from 53 to 160), we ask whether the previously observed high number of cryptic biological species is reflected in high number of genetic groups within each of these three well-defined taxonomic species.
Materials and methods
Sampling
A total of 385 plants from 83 populations representing most of the circumpolar distribution area of Draba fladnizensis, D. nivalis and D. subcapitata were attempted to be analyzed. The number of individuals per population varied from 1 to 10. A few polyploid plants were discovered in the microsatellite analysis (that is, showing more than two alleles at a locus). These plants and plants amplifying poorly were excluded from further analysis, leaving a total of 337 supposedly diploid individuals from 78 populations (Figure 1; Supplementary Table S1). All samples used for AFLP analysis were collected in the field and dried in silica gel. The silica samples and voucher specimens for all of these populations are deposited in the DNA bank at the National Centre for Biosystematics at the Natural History Museum in Oslo. Some additional leaf samples were taken from herbarium specimens deposited in the herbaria in Oslo (O) and Tromsø (TROM), Norway, and analyzed for microsatellites only (Supplementary Table S1).
DNA analyses
DNA was extracted using the DNeasy Plant 96 Kit (Qiagen, Hilden, Germany) following the manufacturer's protocol. Ten previously developed microsatellite loci were analyzed for all samples (Supplementary Table S2, Bell and Ecker, 1994; Skrede et al., 2008b). Nine of these loci have been mapped on a genetic linkage map for D. nivalis, and were evenly distributed throughout the genome (Skrede et al., 2008a). One of the loci (AthSO329; Bell and Ecker, 1994) was not included in the mapping study and was monomorphic in D. nivalis and D. subcapitata and therefore only included in the D. fladnizensis data set (Supplementary Table S2). The primers were amplified following the protocol in Skrede et al. (2008b). The primers DnB227, DnB228, DnA218, DnA222 and AthSO329 were coloaded, and DnA117, DnB123A, DnB207, DnB220 and DnB229 were coloaded, and run on an ABI 3100 DNA Sequencer. The raw data were visualized, manually controlled and analyzed in GENEMAPPER 4.0 (Applied Biosystems, Foster City, CA, USA).
Amplified fragment length polymorphisms analysis was carried out according to Gaudeul et al. (2000), with a few modifications (Alsos et al., 2007). Eighteen primer combinations, with three selective bases for both EcoRI and MseI, were tested for five individuals from different geographic regions of each species. We preferred primer combinations giving clearly separated and polymorphic fragments. The primer combinations 6-FAM-EcoRI-AGA MseI-CTC, VIC-EcoRI-ACG MseI-CAA and NED-EcoRI-ACC MseI-CTT were analyzed for the complete sample set. The three chosen primer combinations were coloaded and analyzed on an ABI 3100 DNA Sequencer using 3.5 μl of selective PCR products (from a mix of 2.0 μl 6-FAM, 2.0 μl VIC, 3.0 μl NED and 14 μl mqH2O), 0.3 μl GeneScan ROX 500 (Applied Biosystems) and 11.7 μl HiDi (formamide) per run. The raw data were visualized, aligned with the internal size standard, manually controlled and analyzed in GENEMAPPER 4.0 (Applied Biosystems). Fragments in the size range 50–500 bp were scored as present (1) or absent (0). Six samples were included in all runs on the ABI 3100 DNA Sequencer to ensure that the runs were comparable. In addition, 27 DNA samples were duplicated and analyzed twice. These 33 samples were used to calculate an error rate according to Bonin et al. (2004).
Data analyses
Eight data sets were produced, including one microsatellite and one AFLP data set for each of the three species, and one combined microsatellite and one combined AFLP data set, both containing all three species.
Genetic structuring was analyzed for each data set using the Bayesian clustering approach implemented in STRUCTURE 2.2 (Pritchard et al., 2000; Falush et al., 2007) and BAPS 3.1 (Corander and Marttinen, 2006). The microsatellite data sets were converted into STRUCTURE format with the software CONVERT (Glaubitz, 2004), and AFLP data sets were converted with the script AFLPdat (Ehrich, 2006). The AFLP data sets were coded with a top row indicating 0 as the recessive allele, making all models in STRUCTURE 2.2 available also for studies using dominant markers (Falush et al., 2007). The analyses were performed on the Bioportal at the University of Oslo, using the admixture model, 106 iterations and a burn-in of 105 iterations. As the admixture model using correlated allele frequencies is the model that most accurately assigns individuals to closely related groups, this was the preferred model (Falush et al., 2003). However, this model tends to overestimate the number of groups (K). Thus, analyses using the admixture model with uncorrelated allele frequencies were also performed (Pritchard et al., 2000). Ten replicates for each K were analyzed for K=1 to K=10. The numbers of groups for each of the three species were chosen after the STRUCTURE output files were analyzed in the software R (R Development Core Team, 2004) by the script Structure.sum (D Ehrich; available from http://www.nhm.uio.no/ncb/). This script visualizes the estimated likelihood of each run, the similarity coefficient between runs (Rosenberg et al., 2002) and Evanno's delta K (Evanno et al., 2005). The best estimate of K was defined by the model giving the highest probability of the data, with a peak in the delta K graph, and that also gave consistent results over multiple runs.
The microsatellite data sets were converted into BAPS format using the software CREATE (Coombs et al., 2008), and the AFLP data sets were converted manually. All data sets were analyzed in BAPS with three replicates for each K value up to K=30.
Principal coordinate analyses (PCO) were used to visualize the genetic structure of each data set. For the microsatellite data sets, the analyses were executed in GENALEX (Peakall and Smouse, 2006) using the distance of Smouse and Peakall (1999). PCO analyses of the AFLP data sets were executed in NTSYS-PC (Rohlf, 1990) using Dice's similarity coefficient.
The number of private alleles was counted for all populations in all data sets, in addition to total number of private alleles in each species. The software HPRARE (Kalinowski, 2006) was used to calculate the number of private alleles (P) and the allelic richness (A) for the microsatellite data sets. As high allelic richness by chance will give more private alleles, we also calculated the ratio of private alleles over allelic richness (P/A) following Muller et al. (2008). As a measure of the distinctiveness of each of the three species, we calculated P/A for the combined microsatellite data set. Private alleles/allelic richness were calculated for each microsatellite data set for all populations with five individuals or more. P/A was also calculated for geographic regions by concatenating neighboring populations. Private markers in the AFLP data set were counted using AFLPdat (Ehrich, 2006). As a measure comparable with P/A for the microsatellite data, we calculated the rarity of AFLP markers using the frequency down-weighted marker index (DW; Schönswetter and Tribsch, 2005) for all AFLP data sets. For each individual, the occurrence of a marker was divided by the total number of occurrences of that marker in the data set. All markers were summed up for each individual and averaged over the population. DW was calculated for all populations with five individuals or more.
Analyses of molecular variance (AMOVA) were carried out in ARLEQUIN version 3.1 (Excoffier et al., 1992). For the microsatellite data sets, the AMOVA were performed on the basis of the number of different alleles, whereas pairwise distances were used for the AFLP data sets.
The expected heterozygosity (HE) at all microsatellite loci was calculated using FSTAT (Nei, 1987; Goudet, 2001). Average HE over all loci was used as a measure of intrapopulation diversity. AFLP intrapopulation variation (D) was calculated in AFLPdat using Nei's gene diversity (Nei, 1987). Intrapopulation diversity was calculated for populations with five individuals or more. Pearson's correlation between HE and D was calculated using R (R Development Core Team, 2004). Analyses of variance were used to test for significant differences in HE and D between species.
Results
The 10 microsatellite loci were successfully analyzed for 316 plants (D. fladnizensis, N=108; D. nivalis, N=128; D. subcapitata, N=95). A total of 192 alleles were detected, giving an average of 19.2 alleles per locus (Supplementary Table S2). Most plants were homozygous, only 5.3, 3.1 and 2.8% heterozygotes were observed in D. fladnizensis, D. nivalis and D. subcapitata, respectively. The mean scoring error calculated from the 48 duplicated samples in the microsatellite analysis was 3.3%. One microsatellite locus was monomorphic in D. nivalis and D. subcapitata. The three AFLP primer combinations provided a total of 160 polymorphic loci in the 271 individuals analyzed successfully (D. fladnizensis, N=104; D. nivalis, N=86; D. subcapitata, N=73). The mean scoring error calculated from the 33 duplicated samples in the AFLP analysis was 1.6%, and markers polymorphic in less than 1.6% of the samples were therefore omitted.
The combined data sets
The PCO and STRUCTURE analyses of the combined AFLP data set and the combined microsatellite data set separated the three species into distinct groups (not shown). In the AMOVA, 47.2 and 30.2% of the variation was partitioned among species in the AFLP and the microsatellite data sets, respectively (Table 1). Each species had 0.25–0.35 private microsatellite alleles/allelic richness (Table 2). Very few AFLP markers were private for individual species (one for D. fladnizensis, five for D. nivalis and none for D. subcapitata). D. subcapitata had lower values for the rarity measures (P/A and DW) and lower genetic diversity than D. fladnizensis and D. nivalis, although not significantly so (P/A=0.28, 0.33 and 0.32, DW=0.55, 0.56 and 0.60, HE=0.193, 0.332 and 0.233, D=0.042, 0.044 and 0.052, for D. subcapitata, D. fladnizensis and D. nivalis, respectively; Table 2).
Draba fladnizensis
For D. fladnizensis, two genetic groups were identified in the STRUCTURE analyses of the microsatellite data set (Supplementary Figure S1), one American and one Eurasian (Figure 1). The same groups were identified on the basis of the AFLP data set for K=2. However, K=4 was chosen as the most appropriate number of AFLP groups (Supplementary Figure S1). For both models and for K=2 and K=4, most individuals were assigned to one specific group by a probability of 0.9 or more, and some populations were mixed. For K=4, the Eurasian group was divided into three groups: one broadly northern, one broadly southern containing the Alpine plants and a subset of the Altai plants and one containing the remaining Altai plants (Figure 1). The Altai group was not found when analyzing the AFLP data set using the uncorrelated allele frequencies model, and these individuals grouped with the broadly southern group (not shown). In the BAPS analyses, 28 microsatellite groups were identified (more than the 26 populations included), but the same four AFLP groups as identified in STRUCTURE were found.
The same main division into American and Eurasian groups was observed in the PCO plots for both the AFLP and microsatellite data sets. In the PCO plot of the AFLP data, the Eurasian plants formed a gradient ranging from the southern alpine ones to the arctic ones along axis 2, similar to the groups recognized by STRUCTURE and BAPS (Figure 1).
The AMOVA gave similar results for the two data sets (Table 1; AFLPs 61.4% and microsatellites 58.3% variation among populations). A total of 20.9% of the microsatellite variation was among the two STRUCTURE/PCO groups and 40.7% was among populations. The four groups found in the AFLP STRUCTURE analyses explained 30.8% of the variation among groups and 33.7% among populations.
The microsatellite private alleles/allelic richness ratio varied from 0.00 to 0.69 (Supplementary Table S1). Most populations from Alaska and eastern Siberia had high values (0.20–0.60), whereas the populations from Svalbard, northwestern Russia and the Alps had low values (0.00–0.20; Figure 2). The same pattern was observed in the analyses for geographic regions (not shown). The AFLP DW values varied from 0.55 to 3.13, but with no clear geographic pattern (Supplementary Table S1, Figure 2).
Number of private alleles over allelic richness (P/A) for 33 Draba populations based on nine (D. nivalis and D. subcapitata) or ten (D. fladnizensis) microsatellite loci, and rarity of AFLP markers expressed as the frequency down-weighted marker index (DW) of 38 Draba populations based on 160 loci. Populations with less than five individuals were omitted.
The intrapopulation diversities ranged from 0.00 to 0.67 and from 0.02 to 0.11 for HE and D, respectively, but with no clear geographic pattern (Supplementary Table S1). The diversities calculated from the AFLP (D) and the microsatellite (HE) data sets were not significantly correlated (Pearson's r=0.71, P>0.05, n=10).
Draba nivalis
For D. nivalis, three genetic groups were identified in the STRUCTURE analyses of the microsatellite data set (Supplementary Figure S1), one broadly Beringian and two largely sympatric amphi-Atlantic ones (Figure 1). For the AFLP data set, K=4 was suggested as the most appropriate number of groups. For both models and for K=3 and K=4, most individuals were assigned to one specific group by a probability of 0.9 or more, and some populations were mixed. These groups corresponded to the groups identified in the microsatellite data set, except that the two populations from Siberia formed a separate group (Figure 1). However, when studying the consistent results for K=3, Siberia, Beringia and Eurasia were identified as separate groups. The uncorrelated allele frequencies model could not distinguish the four groups and shifted between two groups (Siberia versus the rest of the samples) or three groups (Siberia, Beringa and Eurasia; not shown). In the BAPS analyses, 20 microsatellite groups were identified. However, three BAPS groups were recognized in the AFLP data set: Siberia, Beringia and Eurasia.
The three microsatellite STRUCTURE groups and the three AFLP BAPS groups were also detected in the PCO plots (Figure 1).
In the AMOVA, 70.9 and 59.5% of the variation was found among populations based on microsatellites and AFLPs, respectively (Table 1). A total of 27.3% of the variation was found among the three STRUCTURE microsatellite groups and 46.2% was found among populations, and 30.3% of the variation was found among the four STRUCTURE AFLP groups and 33.7% was found among populations.
The microsatellite private alleles/allelic richness ratio varied from 0.00 to 0.54 (Supplementary Table S1). The amphi-Atlantic populations were less distinctive than the Beringian ones (P/A 0.00–0.20 versus 0.20–0.60; Figure 2). The same pattern was observed in the analyses for geographic regions. The AFLP DW values varied from 0.69 to 2.96, but with no clear geographic pattern (Supplementary Table S1, Figure 2).
The intrapopulation diversities ranged from 0.00 to 0.68 and from 0.01 to 0.11 for HE and D, respectively, but with no clear geographic pattern (Supplementary Table S1). The diversities calculated from the AFLP (D) and the microsatellite (HE) data sets were not significantly correlated (Pearson's r=0.29, P>0.05, n=9).
Draba subcapitata
No STRUCTURE groups could be identified in the microsatellite data set for D. subcapitata. For all K=2 to K=10, the replicate runs gave different results and different likelihoods (Supplementary Figure S1). K=5 was suggested as the most appropriate number of groups for the AFLP data set (Figure 1, Supplementary Figure S1), but these were either restricted to a single geographic region (two in east Greenland and one in Siberia) or geographically highly disjunct (Svalbard and Canada, and Siberia and Canada). The uncorrelated allele frequencies model identified two groups: one population in east Greenland versus the rest of the samples. For all models and for K=2 and K=5, most individuals were assigned to one specific group by a probability of 0.9 or more, and some populations were mixed. In the BAPS analyses, nine microsatellite groups were identified. Four BAPS AFLP groups were recognized, corresponding to the five STRUCTURE groups except that one population from east Greenland and the populations from Svalbard were grouped together (orange and green in Figure 1).
Only limited geographic structure was found in the PCO plot of the microsatellite data; plants from some geographic regions spanned most of the plot area (Figure 1, details not shown). Only two of the five AFLP STRUCTURE groups appeared as quite distinct in the PCO plot of the AFLP data (Figure 1).
In the AMOVA, 55.1% of the variation was found among populations for AFLPs and 61.6% for microsatellites (Table 1). A total of 31.9% of the variation was found among the five AFLP STRUCTURE groups and 26.1% of the variation was found among populations.
The microsatellite private alleles/allelic richness ratio varied from 0.00 to 0.36 and the AFLP DW values varied from 0.46 to 2.63, but with no clear geographic pattern (Supplementary Table S1, Figure 2).
The intrapopulation diversities ranged from 0.01 to 0.34 and from 0.03 to 0.06 for HE and D, respectively, but with no geographic pattern (Supplementary Table S1). The diversities calculated from the AFLP (D) and the microsatellite (HE) data sets were not significantly correlated (Pearson's r=−0.73, P>0.05, n=6).
Discussion
The three Draba species were clearly separated in our analyses, in accordance with their traditional recognition as well-defined taxonomic species, even though they had none or only few diagnostic or private alleles at microsatellite and AFLP loci. Thus, we conclude that they represent distinct, but closely related, taxa. Our findings thus corroborate the previous phylogeny of the group inferred from RAPD fingerprinting and ribosomal DNA (ITS) sequencing (Grundt et al., 2004).
Some cautionary notes, because of the sampling strategy we have chosen and because of the primarily selfing mating system of these species, are necessary before the results of this study are further interpreted. We decided to analyze many but small populations to cover the ranges of these species because our primary aim was to investigate broad-scale genetic structuring. This choice was justified by the previous experimental study showing frequent spontaneous self-pollination (Brochmann, 1993). Indeed, as expected for mainly selfing species (Hamrick and Godt, 1996), we observed low within-population diversity and large between-population differentiation in all three species. Most of the individuals analyzed were homozygous at the microsatellite loci, and the populations were not in Hardy–Weinberg equilibrium. The usual advantage of using codominant markers such as microsatellites compared with dominant ones such as AFLPs is therefore reduced. Most importantly, genetic structure analyses such as STRUCTURE and BAPS may be influenced by deviations from Hardy–Weinberg equilibrium and lead to overestimation of the number of groups (Pritchard et al., 2000; Falush et al., 2003; Corander and Marttinen, 2006).
High-arctic versus arctic–alpine distributions
We found less geographic structuring and less genetic diversity in the hardiest, high-arctic species, D. subcapitata, than in the two less hardy, more southerly distributed species. High dispersal potential and recent bottlenecks have been suggested as an explanation for the low levels of genetic structuring in high-arctic animal species (Paetkau et al., 1999; Dalén et al., 2005; Marthinsen et al., 2008). In arctic plants, much higher levels of long-distance dispersal than traditionally thought have recently been demonstrated for several species, probably facilitated by wind over frozen sea ice, by driftwood or by birds (Alsos et al., 2007). This study suggested that establishment rather than dispersal per se was the limiting factor for plant colonization in the Arctic Archipelago of Svalbard; the hardiest species had colonized more frequently than the less hardy ones, regardless of morphological adaptations to long-distance dispersal. Notably, among the three species studied here, Draba subcapitata extends into much harsher and more northern environments (for example, the polar deserts along the shores of the Arctic Ocean). Thus, it is reasonable to believe that this species may have higher potential for establishment after dispersal under high-arctic conditions than the less hardy D. nivalis and D. fladnizensis. This could explain that no genetic groups were observed at microsatellite loci in this species, and that it had lower levels of geographic structuring at AFLP loci than the two other species. In addition, it is possible that long-distance dispersal in D. subcapitata is facilitated by its higher population densities at high latitudes, resulting in more seeds available for chance dispersal across the Arctic basin. This can account for the occurrence of genetically similar but geographically very disjunct populations in D. subcapitata.
The lower level of geographic structuring as well as the lower genetic diversity in Draba subcapitata is consistent with the hypothesis that the hardiest species experienced the heaviest bottlenecks during interglacial. In the arctic fox, large population expansion after the previous interglacial was inferred from molecular data, suggesting that such hardy species go through bottlenecks during interglacials rather than glacials (Dalén et al., 2005). Thus, D. subcapitata may at present have a marginal distribution and was probably more restricted, especially during the postglacial warm period (9500–4000 years BP; cf. Birks et al., 1994), than D. nivalis and D. fladnizensis.
The two less hardy species, D. fladnizensis and D. nivalis, show more distinct geographic structuring both at microsatellite and AFLP loci, with a similar main split between the Beringian and the Eurasian/North Atlantic populations. These two groups of populations probably survived in different refugia during several glaciations. The main pattern found in these two Draba species is similar to that observed in many other circumpolar and not very hardy arctic–alpine plant species, for example, Rubus chameomourus, Epilobium angustifolium, Juniperis communis, Arctous alpinus, C. tetragona, Betula nana and V. uliginosum (Eidesen, 2007; Eidesen et al., 2007a, 2007b; Ehrich et al., 2008).
In D. fladnizensis, we also identified two main subgroups in the Eurasian/North Atlantic area on the basis of the AFLP data, one arctic and one alpine, suggesting distinct refugia at least during the last glaciation (east and south of the ice). This pattern is similar to that in Dryas octopetala, Vaccinium vitis-idea and B. nana (Skrede et al., 2006; Eidesen, 2007; Eidesen et al., 2007a). A recent history of these two subgroups is suggested by their low levels of private microsatellite alleles (Figure 2).
In D. nivalis, the extremely divergent group observed in Siberia (Yakutia) based on the AFLP data was surprisingly not reflected in the microsatellite data. This discrepancy may be explained by the more extensive coverage of the genome by AFLP analysis. The Yakutia populations are also somewhat distinct in morphology, and it is possible that they should be recognized as a separate taxon. Notably, tetraploid D. nivalis-resembling plants have been reported previously from the same area (as Draba sp. by Grundt et al., 2005); however, we found no evidence for tetraploidy in the plants analyzed here.
AFLPs versus microsatellites
We found more distinct genetic structuring in these three circumpolar Draba species at the AFLP loci than at the microsatellite loci (Figure 1), possibly because of the higher number of loci surveyed (160 versus 10) in these selfing species, and/or because structuring at very large geographic scales is blurred by high mutation rate leading to homoplasy at microsatellite loci. AFLPs also separated the three species more distinctly, both in the PCO (not shown) and the AMOVA (Table 1). Thus, for large-scale intraspecific studies and for interspecific studies, AFLP may be more appropriate as molecular marker (cf. Mariette et al., 2001; Gaudeul et al., 2004; Woodhead et al., 2005; Sønstebø et al., 2007).
The BAPS analyses of the microsatellite data invariably resulted in high number of groups, in some cases even more groups than the number of populations analyzed. We conclude that BAPS is not suited for analysis of microsatellite data in these selfing species, probably because it is too heavily affected by the deviations from Hardy–Weinberg equilibrium. We found, however, more reasonable and similar results from STRUCTURE analyses of the microsatellite data and STRUCTURE as well as BAPS analyses of the AFLP data, suggesting that these analyses were less affected by the deviations from Hardy–Weinberg equilibrium.
Somewhat surprisingly, the intrapopulation genetic diversity measures based on AFLP and microsatellites (D and HE, respectively; Supplementary Table S1) were not correlated, and we also obtained quite different rarity measures for the two types of markers (DW and P/A, respectively; Figure 2; Supplementary Table S1). Higher intrapopulation genetic diversity could be expected for microsatellites because of their higher mutation rate (Mariette et al., 2001; Gaudeul et al., 2004), but the different measures should be correlated. However, our sampling sizes were small, and selfing populations may show variable genetic diversity depending on the number of lineages sampled. The different and partly lack of geographic pattern for the rarity measures (Figure 2) may also reflect small sample sizes as well as predominant selfing.
Genetic groups and cryptic species
We observed very few genetic groups within these three taxonomic species of Draba, certainly too few to reflect the high number of cryptic biological species observed previously within each of them (Grundt et al., 2006). This result is consistent with the AFLP analysis of Grundt et al. (2006), even if we extended the number of AFLP loci from 53 to 160 and also analyzed microsatellites. Thus, our results lend additional support to the hypothesis that the development of sterility barriers occurs very rapidly in this group and only involve small parts of the genome (Grundt et al., 2006; Skrede et al., 2008a). We have recently shown that there are several genetic mechanisms involved in the development of fertility barriers (Skrede et al., 2008a). Both cytonuclear and nuclear–nuclear interactions, in addition to chromosomal translocations, are probably involved. We nevertheless observed only a few quantitative trait loci affecting fertility (Skrede et al., 2008a), and it is therefore not surprising that the differentiation into cryptic biological species is not yet detectable in overall genomic differentiation as explored in this study. The two sympatric genetic groups we detected in European D. nivalis based on microsatellites could correspond to different biological species, but this pattern was not found in D. fladnizensis, even if this species contains several cryptic species in Europe as well (Brochmann et al., 1993; Grundt et al., 2006).
Conclusion
Very little geographic structuring was found in the high-arctic species, D. subcapitata, suggesting that this very hardy species may have high dispersal potential and establish easily after long-distance dispersal under high-arctic conditions. A recent bottleneck caused by postglacial warming offers an explanation for the low levels of genetic structuring and diversity found in this species. The two less hardy and more southerly distributed species, D. fladnizensis and D. nivalis, had more genetic diversity and more distinct geographic structuring, suggesting less effective dispersal and less severe postglacial bottlenecking. In these two species, the main split observed between Beringian and Eurasian/North Atlantic plants probably reflects survival in different glacial refugia and is consistent with the pattern observed in other arctic–alpine plants. Our results suggest that AFLPs are better suited for large-scale phylogeographic studies than microsatellites, as microsatellites may evolve too fast and homoplasy may blur the main structure. The number of genetic groups we detected was insignificant compared with the numerous cryptic biological species reported previously within these species, supporting the hypothesis that development of sterility barriers occurs very rapidly in this group and involves only small parts of the genome. The genetic changes affecting fertility were thus not recognized by our multilocus approaches; more time is evidently needed for more differences to accumulate.
References
Abbott RJ, Comes HP (2004). Evolution in the Arctic: a phylogeographic analysis of the circumarctic plant, Saxifraga oppositifolia (Purple saxifrage). New Phytol 161: 211–224.
Abbott RJ, Smith LC, Milne RI, Crawford RMM, Wolff K, Balfour J (2000). Molecular analysis of plant migration and refugia in the Arctic. Science 289: 1343–1346.
Al-Shehbaz IA (1987). The genera of Alysseae (Cruciferae; Brassicaceae) in the southeastern United States. J Arnold Arbor 68: 185–240.
Alsos IG, Eidesen PB, Ehrich D, Skrede I, Westergaard K, Jacobsen GH et al. (2007). Frequent long-distance plant colonization in the changing Arctic. Science 316: 1606–1609.
Alsos IG, Engelskjøn T, Gielly L, Taberlet P, Brochmann C (2005). Impact of ice ages on circumpolar molecular diversity: insights from an ecological key species. Mol Ecol 14: 2739–2753.
Bell CJ, Ecker JR (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19: 137–144.
Birks HH, Paus A, Svendsen JI, Alm T, Mangerud J, Landvik JY (1994). Late Weichselian environmental change in Norway, including Svalbard. J Quat Science 9: 133–145.
Bonin A, Bellemain E, Eidesen PB, Pompanon F, Brochmann C, Taberlet P (2004). How to trace and assess genotyping errors in population genetics studies. Mol Ecol 13: 3261–3273.
Brochmann C (1993). Reproductive strategies of diploid and polyploid populations of arctic Draba (Brassicaceae). Plant Syst Evol 185: 55–83.
Brochmann C, Borgen L, Stedje B (1993). Crossing relationships and chromosome-numbers of Nordic populations of Draba (Brassicaceae), with emphasis on the D. alpina complex. Nord J Bot 13: 121–147.
Coombs JA, Letcher BH, Nislow KH (2008). CREATE: software to create input files from diploid genotypic data for 52 genetic software programs. Mol Ecol Resour 8: 578–580.
Corander J, Marttinen P (2006). Bayesian identification of admixture events using multilocus molecular markers. Mol Ecol 15: 2833–2843.
Dalén L, Fuglei E, Hersteinsson P, Kapel CMO, Roth JD, Samelius G et al. (2005). Population history and genetic structure of a circumpolar species: the arctic fox. Biol J Linn Soc 84: 79–89.
Ehrich D (2006). AFLPDAT: a collection of R functions for convenient handling of AFLP data. Mol Ecol Notes 6: 603–604.
Ehrich D, Alsos IG, Brochmann C (2008). Where did the northern peatland species survive the dry glacials: cloudberry (Rubus chamaemorus) as an example. J Biogeogr 35: 801–814.
Eidesen PB (2007). Arctic–alpine plants on the move: individual and comparative phylogeographies reveal responses to climate change. PhD thesis, University of Oslo.
Eidesen PB, Alsos IG, Popp M, Stensrud Ø, Suda J, Brochmann C (2007a). Nuclear vs plastid data: complex Pleistocene history of a circumpolar key species. Mol Ecol 16: 3902–3925.
Eidesen PB, Carlsen T, Molau U, Brochmann C (2007b). Repeatedly out of Beringia: Cassiope tetragona embraces the Arctic. J Biogeogr 34: 1559–1574.
Evanno G, Regnaut S, Goudet J (2005). Detecting the number of clusters of individuals using the software Structure: a simulation study. Mol Ecol 14: 2611–2620.
Excoffier L, Smouse PE, Quattro JM (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes—application to human mitochondrial-DNA restriction data. Genetics 131: 479–491.
Falush D, Stephens M, Pritchard JK (2003). Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587.
Falush D, Stephens M, Pritchard JK (2007). Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7: 574–578.
Federov VB, Goropashnaya AV, Jaarola M, Cook A (2003). Phylogeography of lemmings (Lemmus): no evidence for postglacial colonization of Arctic from the Beringian refugium. Mol Ecol 12: 725–731.
Gaudeul M, Taberlet P, Till-Bottraud I (2000). Genetic diversity in an endangered alpine plant, Eryngium alpinum L. (Apiaceae) inferred from amplified fragment length polymorphism markers. Mol Ecol 9: 1625–1637.
Gaudeul M, Till-Bottraud I, Barjon F, Manel S (2004). Genetic diversity and differentiation in Eryngium alpinum L. (Apiaceae): comparison of AFLP and microsatellite markers. Heredity 92: 508–518.
Glaubitz J (2004). Convert: a user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol Ecol Notes 4: 309–310.
Goudet J (2001). Fstat, a program to estimate and test gene diversities and fixation indices. (http://www2.unil.ch/popgen/softwares/fstat.htm)
Grundt HH (2003). The arctic–alpine polyploid Draba lactea and its low-ploid relatives—evolution and taxonomy. Dr Scient thesis, University of Oslo.
Grundt HH, Kjølner S, Borgen L, Rieseberg LH, Brochmann C (2006). High biological species diversity in the arctic flora. Proc Natl Acad Sci 103: 972–975.
Grundt HH, Obermayer R, Borgen L (2005). Ploidal levels in the arctic–alpine polyploid Draba lactea (Brassicaceae) and its low-ploid relatives. Bot J Linn Soc 147: 333–347.
Grundt HH, Popp M, Brochmann C, Oxelman B (2004). Polyploid origins in a circumpolar complex in Draba (Brassicaceae) inferred from cloned nuclear DNA sequences and fingerprints. Mol Phylogenet Evol 32: 695–710.
Hamrick JL, Godt JW (1996). Effects of life history traits on genetic diversity in plant species. Philos Trans R Soc Lond B Biol Sci 351: 1291–1298.
Hultén E, Fries M (1986). Atlas of North European Vascular Plants North of the Tropic of Cancer 1–3. Koeltz Scientific Books: Koenigstein.
Kalinowski S (2006). HP-Rare: a computer program for performing rarefaction on measures of allelic diversity. Mol Ecol Notes 5: 187–189.
Mariette S, Chagné D, Lézier C, Pastuszka P, Raffin A, Plomion C et al. (2001). Genetic diversity within and among Pinus pinaster populations: comparison between AFLP and microsatellite markers. Heredity 86: 469–479.
Marthinsen G, Wennerberg L, Solheim R, Lifjell JT (2008). No phylogeographic structure in the circumpolar snowy owl (Bubo scandiacus). Conserv Genet (in press; doi:10.1007/s10592-008-9581-6).
Muller M-H, Leppälä J, Savolainen O (2008). Genome-wide effects of postglacial colonization in Arabidopsis lyrata. Heredity 100: 47–58.
Mulligan GA (1974). Cytotaxonomic studies of Draba nivalis and its close allies of Canada and Alaska. Can J Bot 52: 1793–1801.
Nei M (1987). Molecular Evolutionary Genetics. Columbia University Press: New York.
Paetkau D, Amstrup SC, Born EM, Calvert W, Derocher AE, Garner GW et al. (1999). Genetic structure of the world's polar bear populations. Mol Ecol 8: 1571–1584.
Peakall R, Smouse P (2006). Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6: 288–295.
Pritchard JK, Stephens M, Donnelly P (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959.
R Development Core Team (2004). A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna.
Rohlf F (1990). NTSYS-PC. Numerical Taxonomy and Multivariate Analysis System Version 2.02 edn. Exeter Software: Setauket, NY.
Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA et al. (2002). Genetic structure of human populations. Science 298: 2381–2385.
Schönswetter P, Tribsch A (2005). Vicariance and dispersal in the alpine perennial Bupleurum stellatum L. (Apiaceae). Taxon 54: 725–732.
Skrede I (2008). Speciation, divergence, and dispersal in arctic diploid plants. PhD thesis, University of Oslo.
Skrede I, Brochmann C, Borgen L, Rieseberg LH (2008a). Genetics of intrinsic postzygotic isolation in a circumpolar plant species, Draba nivalis (Brassicaceae). Evolution 62: 1840–1851.
Skrede I, Carlsen T, Rieseberg LH, Brochmann C (2008b). Microsatellites for three distantly related genera in the Brassicaceae. Conserv Genet (in press; doi:10.1007/s10592-008-9598-x).
Skrede I, Eidesen PB, Piñeiro R, Brochmann C (2006). Refugia, differentiation and postglacial migration in arctic–alpine Eurasia, exemplified by the mountain avens (Dryas octopetala L.). Mol Ecol 15: 1827–1840.
Smouse P, Peakall R (1999). Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity 82: 561–573.
Stewart JR, Dalén L (2008). Is the glacial refugium concept relevant for northern species? A comment on Pruett and Winker 2005. Clim Chang 86: 19–22.
Sønstebø JH, Borgstrøm R, Heun M (2007). A comparison of AFLPs and microsatellites to identify the population structure of brown trout (Salmo trutta L.) populations from Hardangervidda, Norway. Mol Ecol 16: 1427–1438.
Weider LJ, Hobæk A, Colbourne JK, Crease TJ, Dufresne F, Herbert PDN (1999). Holarctic phylogeography of an asexual species complex—I. Mitochondrial DNA variation in arctic Daphnia. Evolution 53: 777–792.
Wennerberg L (2001). Breeding origin and migration pattern of dunlin (Calidris alpina) revealed by mitochondrial DNA analysis. Mol Ecol 10: 1111–1120.
Woodhead M, Russell J, Squirrell J, Hollingsworth M, Mackenzie K, Gibby M et al. (2005). Comparative analysis of population genetic structure in Athyrium distentifolium (Pteridophyta) using AFLPs and SSRs from anonymous and transcribed gene regions. Mol Ecol 14: 1681–1695.
Acknowledgements
We thank LG Kvernstuen for producing the AFLP data sets; JH Sønstebø, PB Eidesen, D Ehrich, R Elven and MM Tollefsrud for discussions; three anonymous referees for valuable comments; and the collectors (cf. Supplementary Table S1) for providing samples. This project was funded by the Strategic University Program Grant 146515/420 from the Research Council of Norway to C Brochmann. Additional funding was obtained from the Natural History Museum, University of Oslo.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary information
Rights and permissions
About this article
Cite this article
Skrede, I., Borgen, L. & Brochmann, C. Genetic structuring in three closely related circumpolar plant species: AFLP versus microsatellite markers and high-arctic versus arctic–alpine distributions. Heredity 102, 293–302 (2009). https://doi.org/10.1038/hdy.2008.120
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/hdy.2008.120
Keywords
This article is cited by
-
Genetic structure of a widespread alpine shrub Rhododendron aureum (Ericaceae) across East Asia
Journal of Plant Research (2021)
-
Genetic diversity and population structure in Cary’s Beardtongue Penstemon caryi (Plantaginaceae), a rare plant endemic to the eastern Rocky Mountains of Wyoming and Montana
Conservation Genetics (2019)
-
The phylogeographic structure of Arabis alpina in the Alps shows consistent patterns across different types of molecular markers and geographic scales
Alpine Botany (2018)
-
AFLP diversity and spatial structure of Calycophyllum candidissimum (Rubiaceae), a dominant tree species of Nicaragua’s critically endangered seasonally dry forest
Heredity (2017)
-
Genetic and geographical structure of boreal plants in their southern range: phylogeography of Hippuris vulgaris in China
BMC Evolutionary Biology (2016)