Abstract
Vascular endothelial growth factor receptor 1 (VEGFR-1) has been implicated in diverse pathologies, including cancers. Although VEGFR-1 is considered as functionally impaired kinase, its decoy characteristics make it an important regulator of VEGFR-mediated signaling, particularly in tumor angiogenesis. VEGFR-1 conveys signaling via its tyrosine kinase (TK) domain whose activation is regulated by phosphorylation of specific tyrosine residues. Thus dysregulation of VEGFR-1 signaling, as reported in most of the cancers, might be a consequence of altered phosphorylation that could be attributed to genotypic variations in its TK domain. Considering the importance of TK domain of VEGFR-1, we carried out its mutational screening in 84 clinically validated and histopathologically confirmed colorectal cancer patients. By means of direct DNA sequencing and SNP analyses, eight novel variations, including one synonymous, two deletion, one missense and four intronic variations, were reported in the TK domain of VEGFR-1. rs730882263:C>G variation specifically reported in colon cancer, representing a single-atomic change (Sulfur to Oxygen) in the predicted (p.Cys1110Ser) protein, was observed as potentially deleterious variation as assessed by multiple single-nucleotide polymorphism prediction servers. Molecular dynamics simulations of VEGFR-1 Wt and (p.Cys1110Ser) variant models revealed major conformational changes in variant protein presumptuously generating an open conformation thereby exposing the activation domain and consequently increasing the probability of phosphorylation events: a condition frequently reported in cancers.
Similar content being viewed by others
Introduction
Vascular endothelial growth factor receptor 1 (VEGFR-1; Fms-related tyrosine kinase-1), primarily associated with neovascularization, is an important factor in tumor angiogenesis1, 2, 3 and is considered as an effective and safest target4, 5 in innovative antitumor strategies. The weak tyrosine kinase activity of VEGFR-1 and its high affinity for VEGFR ligands limit VEGF-mediated angiogenesis;6 however, substantial evidences support the view that the activation of VEGFR-1 under pathological conditions results in the amplification of angiogenesis mediated by VEGFR-2.7, 8, 9, 10 Such differential effects of VEGFR-1 on VEGF-mediated angiogenesis are still less understood and could be attributed to structural or epigenetic variations in VEGFR-1.
VEGFR-1 (Flt-1) belongs to receptor tyrosine kinase family of growth factors and mediates its effects via phosphorylation of its specific tyrosine residues in the protein kinase domain, namely, Y1048, Y1053, Y1169, Y1213, Y1242, Y1327 and Y1333, via autocatalysis after receptor dimerization, thus generating docking sites for Src homology 2 (SH2) domain- and phosphotyrosine binding (PTB) domain-containing proteins, which convey further downstream signaling.11 The patterns of phosphorylation of tyrosine residues in the intracellular domain of VEGFR-1 determine the binding of adaptor proteins and hence the specific activation of signaling molecules and the processes involved thereof. For example: Phosphorylation at Tyr-1169 is important for interaction with PLCG,12 phosphorylation at Tyr-1213 is important for interaction with PIK3R1, PTPN11, GRB2, and PLCG,13 while phosphorylation at Tyr-1333 is important for endocytosis and for interaction with CBL, NCK1 and CRK.14 Thus it is evident that TK domain has a major role in modulating VEGFR-1-mediated signaling pathways and any genotypic aberration will have a definite impact on epigenetic gene regulation and thus cancerous phenotypes. Considering the importance TK domain of VEGFR-1/FLT-1 gene, we carried out its mutational screening in colorectal cancer patients from selected cohorts of Kashmir valley.
Materials and methods
Clinical subjects
We conducted a case–control study within three regionally distinct cohorts, namely, Central, South and North Kashmir, divided into two representative groups of colon cancer and rectal cancer, respectively. Together, all the cohorts included 84 histopathologically confirmed patients with adenocarcinoma of the colon/rectum who were registered in the study for gene mutation analysis. The mean age of patients ranged from 45 to 55 years. All patients signed informed consent confirming their willingness to participate in the study. The protocols/experiments involving the use of human specimens were duly examined and approved by Institutional Ethics Committee, Sher-i-Kashmir Institute of Medical Sciences, Srinagar and Institutional Review Committee, University of Kashmir, Srinagar and were in accordance with the declaration of Helsinki. All incident cases were examined by specialized imaging procedures, namely, proctosigmoidoscopy, colonoscopy, digital rectal examination and contrast enhanced computed tomography besides histopathological confirmation (Supplementary Table S1) to confirm disease status. Patients with squamous cell carcinoma or those who had received chemotherapy or radiotherapy were excluded.
Mutational profiling
Mutational profiling of VEGFR-1 gene was performed on patients catalogued into two distinct groups based on their clinical diagnosis as: (a) patients with cancer of colon; and (b) patients with rectal cancers. For all the clinical subjects, DNA was extracted from cancerous lesion and corresponding adjacent control tissue. Exons (17–26) spanning the putative tyrosine kinase domain of VEGFR-1 were screened for variations using gene-specific primer combinations (Supplementary Table S2). Samples were genotyped by PCR using high-fidelity Taq Polymerase (Sigma Inc., St Louis, MO, USA). PCR was carried out in a final volume of 50 μl, containing 10 ng genomic DNA, 1 × PCR buffer, 0.02 μm deoxynucleotide triphosphates, 0.03 mM MgCl2, 0.04 μM of each primer and 1 U Taq DNA polymerase. The percentage of successful genotyping was 97% for all genetic variants. All the variations were confirmed at least twice by double-pass sequencing.
VEGFR-1 variants
All the genomic variants reported in the study were archived in GeneBank (www.ncbi.nlm.nih.gov/genbank), dbSNP (www.ncbi.nlm.nih.gov/SNP) and ClinVar (http://www.ncbi.nlm.nih.gov/SNP) databases using genome reference assembly GRCh38.p2 against VEGFR-1 reference sequence (NG_012003.1) as template. Supplementary Table S3 provides summary of variations reported in the study.
In silico SNP prediction
To evaluate the functional impact of the amino-acid alterations, four widely used algorithms were used to perform in silico analyses, namely, Protein Variation Effect Analyzer (PROVEAN),15, 16 Sorting Intolerant From Tolerant (SIFT),17 Polymorphism phenotyping-2 (PolyPhen-2)18 and Mutational T@ster.19 The variations were compared across the global SNP submissions reported for VEGFR-1.
Statistical analysis
The outcome of the clinicopathological variables were summarized as percentage of patients bearing the symptom/state within the specified cohort/subtype and compared across all the samples. The clinical associations of SNPs were tested using Fisher's exact test. Strength of association was determined by OR at 95% confidence interval (CI). Statistical significance was set at P<0.05.
Molecular dynamics (MD) simulation
The crystallographic structure of the kinase domain of VEGFR-1 bearing PDB ID: 3HNG and that of VEGFR-2 PDB ID: 3VHE was used in the study. SPDB viewer was used to search for missing atoms and performing energy minimization. The modifications of PDB structure were brought by Accelrys Discovery Studio 4.1.20 To predict the effect of Cys1110Ser variation on structural dynamics of the kinase domain of VEGFR-1, MD simulations using GROMACS 4.5.2 (Hess et al21) platform were performed. The wild-type and variant structures underwent two independent simulations for 50 ns each. MD runs under physiological salt concentration (150 mm) were carried out in neutral environment of TIP3P water model. The equilibration using conjugate gradient and steepest descent energy minimizations were carried out for 100 ps. The trajectories generated over the 50 ns were further analyzed using GROMACS inbuilt tools. All the parameters used for the MD runs are as described in Bukhari et al.22
Molecular docking simulation
Molecular docking simulations were brought about by AutoDock 4.2 tool.23 ATP and ADP were alternatively used to evaluate the nucleotide-binding mode at the catalytic pocket of VEGFR-1 Wt, (p.Cys1110Ser) variant and VEGFR-2 in a grid of 10 Å each in x, y and z direction and centered at ATP-binding site. The docking energy was obtained from the summation of van der Waals energy and hydrogen bonding energy, while binding energy was built up from van der Waals energy and desolvation energy. Lamarckian Genetic Algorithm (GA) was considered for the run and for each ligand 10 GA runs, with 27 000 maximum generations, 0.02 rate of gene mutation and 0.8 as rate of crossover were set. A grid of 10 Å in x, y and z direction was built centered around ATP-binding site.
Results
Clinicopathological analysis
Analysis of clinicopathological data from 50 colon and 34 rectal cancer cases distributed over 3 regional cohorts showed higher male:female ratio for colon cancer, except in Central Kashmir, which showed prevalence of rectal cancers among males (Supplementary Table S1). Significant correlation of colon/rectal cancers with typical symptoms, namely, abdominal pain, rectal bleeding, bowel disturbances and anemia was also established. Histologically, majority of cases, in both colon and rectal cancers, were either well or moderately differentiated (Supplementary Table S1).
Mutational screening
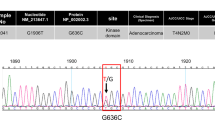
From the mutational scanning of 10 exons (and their corresponding flanking introns), a total of 10 genotypic variations were observed with allelic frequencies of genotypes ranging from 2.5 to 97.5%, consisting of 8 novel variations (1 synonymous, 2 deletion, 1 missense and 4 intronic) and 2 known variations (synonymous) (Table 1). Analysis of global SNP database for VEGFR-1 variants placed rs730882263:C>G [hg38.chr13:g.28891692C>G] among the most deleterious variation (the variations causing protein truncations were excluded) reported so far (Supplementary Table S4). The overall association between rs730882263:C>G VEGFR-1 catalytic domain variation and colon cancer was found to be novel and significant (P<0.001; Table 1,Supplementary Table S3).
Structural and conformational dynamics of VEGFR-1 (p.Cys1110Ser) variant
Explicit water-based simulation under 0.15 nm concentration was performed on the VEGFR-1 Wt and (p.Cys1110Ser) variant. The energy-minimized structures were subjected to two independent 50 ns runs under GROMOS96 43a2 force field and the trajectories were saved after 2 ps, (Supplementary Video S1: Animation). Supplementary Figure S1 shows the structures extracted from both Wt and (p.Cys1110Ser) variant trajectories capturing the structure at 10-ns time interval. The root mean square deviation (RMSD) of the two structures was calculated over time. Both the runs showed a stable run over the period of 50 ns with maximum fluctuations observed at ~15 and ~30 ns in variant structure (red) (Figure 1a). RMSD values for Wt ranged from 1.0 to 0.50 nm while that of variant ranged from 0.8 to 0.45 nm, respectively. The root mean square fluctuation (RMSF) of the amino acids were calculated and plotted, the important amino acids were marked in the plot (Figure 1b), and the change in the RMSF can be observed between the VEGFR-1 Wt and (p.Cys1110Ser) variant at important phosphotyrosine sites, alluding to the structural dynamic implication by CYS to SER variation. The hydrogen bond formation of both the structures was calculated using g_hbond tool with a grid search of 19 × 19 × 19 Å3 and the cutoff value was set at 0.35 Å. Inter- and intra-hydrogen bond patterns of both the structures were plotted (Supplementary Figure S2) as a function of time. Wt VEGFR-1 formed 208 average number of intra-hydrogen bonds per frame and its inter-hydrogen bond number was recorded 501 per frame. In the variant, we observe a decrease in the average intra-hydrogen bond to 204 per frame, leading to increase in the surface area and radius of gyration of the variant structure (Figure 2), which was corroborated by the increase in average number of inter-hydrogen bonding (515 per frame). Thus it can be assumed that (p.Cys1110Ser) variation alters conformation of the amino-acid chain in such a way that it results in an expanded structure of VEGFR-1.
MD simulations of wild-type (black) and variant (red) structures. (a) RMSD plot depicting time points having maximal variation in structures between wild-type and variant structures captured during protein simulation at (I) 13.462, (II) 15.694, (III) 24.730 and (IV) 47.700 ns. (b) Comparative RMSF plot showing fluctuations in wild-type (black) and variant (red) structures along the protein stretch together with their predicted secondary structures. Functionally important positions are highlighted in color. A full color version of this figure is available at the European Journal of Human Genetics journal online.
The psi–phi distribution of the selected important amino acids were also captured over 50-ns run and plotted (Supplementary Figure S3a), the distributions were studied using Gibbs free energy and its plot (Supplementary Figure S3b) was showing the maximum distribution of the selected amino acids.
Long-range conformational change in the structure affects the ATP–ADP transition kinetics
In order to study functional consequences of Cys1110Ser variation, comparative MD simulations were setup between VEGFR-1 Wt, (p.Cys1110Ser) variant and VEGFR-2. RMSD calculations were employed on the three trajectories (Supplementary Figure S4), black represents VEGFR-1, red represents (p.Cys1110Ser) variant and green represents VEGFR-2. Relative change in orientation and motion of amino acids at tRMSD(Start) and tRMSD (max) were observed and are shown in Supplementary Table S5. The allosteric effect of Cys1110Ser variation on VEGFR-1 was evaluated on the basis of relative conformation of conserved amino acids in the ATP-binding pocket, namely, K of K/D/D, αC Glutamate, Gly in the glycine-rich loop, activation segment tyrosines and compared across VEGFR-1 and -2 (Figure 3). As observed by MD simulation, (p.Cys1110Ser) variant favors an open conformation identical to that of activated VEGFR-2 (Supplementary Figure S5), thus pointing toward allosteric activation of tyrosine kinase activity of VEGFR-1.
These findings were further corroborated by binding energy pattern of VEGF receptors during ATP–ADP transition, an invariably important aspect of tyrosine phosphorylation. Figure 4 shows the binding mode and Gibbs free energy (ΔG) of ATP and ADP in complex with the three structures. As shown in Figure 4, Wt VEGFR-1 depicted similar binding affinities for either ATP or ADP ligand, while (p.Cys1110Ser) variant showed relatively higher binding affinity toward ATP compared with ADP, identical to the trend shown by VEGFR-2. The variation in binding affinities of ATP/ADP with Wt VEGFR-1, (p.Cys1110Ser) variant and VEGFR-2 are indicative of an enhanced phosphorylation capability of VEGF receptors and in a way support oncogenic nature of Cys1110Ser variation.
Molecular docking simulation with ATP/ADP at binding sites in the catalytic pocket of VEGF receptors showing differential binding energy pattern in tyrosine phosphorylation events*. (*Based upon ligand docking with ATP or ADP; VEGFR-2 has been shown for comparison). A full color version of this figure is available at the European Journal of Human Genetics journal online.
Discussion
A vast body of literature suggests that the genomic alterations figure in as causative sources of many diseases, particularly cancers. These genomic alterations could range from insertions, deletions, duplications or rearrangements to the most typical single-nucleotide substitutions (SNPs).24 SNPs within a gene may have serious implications on protein function and stability and could guide the progression of cancers or define response to drug treatment.25 Colorectal cancer has been reported to have the most frequently mutated genotype.26 Colorectal tissue homeostasis is chiefly regulated by developmental genes and growth factor receptor molecules. VEGFRs are among these critical molecules that regulate tumor progression and thus genetic defects in these receptors may have serious implications in cancer progression and survival. Among these receptors, VEGFR-1 has been shown to be actively involved in tumor growth and metastasis27 and has an important role in regulation of tumor angiogenesis that is considered to be one of the hallmark of tumor progression. A study by Mimori et al28 showed that VEGFR-1 facilitates establishment of metastases in gastric cancer while another study by Kosaka et al29 found that metastatic cells in peripheral blood show higher expression of VEGFR-1 than non-metastatic cells. These finding again corroborate pivotal role played by VEGFR-1 in cancer establishment and metastasis. A study by Slattery et al30 found significant association between genetic variants in FLT-1 with colon/rectal cancer development in a case–control study involving 2309 cancer cases and 2915 controls signifying the importance of VEGFR-1 in etiology of colorectal cancer. We performed mutational screening of tyrosine kinase domain of VEGFR-1 in 84 colorectal cancer patients following their disease validation in clinical settings (Supplementary Table S1). As depicted in Table 1, we report a total of 10 genotypic variations out of which 8 variations are novel. For studying functional implication of any genotypic variation, its expression at the protein level is warranted. This prompted us to focus on a single-nucleotide variation (rs730882263: C>G) essentially a mis-sense variation generating a predictive variant VEGFR-1 protein harboring Cys1110Ser variation (representing a single-atomic change, namely, Sulfur to Oxygen). Correlation of clinicopathological reports with the SNP data of TK domain of VEGFR-1 showed a significant association of rs730882263: C>G with colon cancer in all the three cohorts analyzed (Supplementary Table S3). It is believed that most of the variations, irrespective of their involvement in disease, destabilize protein structure and thus alter their function. A finer analysis of MD trajectories of Wt and Mu VEGFR-1 studied over 50 ns highlighted prominent changes arising due to variation leading to change in RMSD, RMSF, SASA and Rg thus causing expansion in the structure of the Mu, which could be attributed to altered H-bonding (Figure 2).
An atomic insight of the (p.Cys1110Ser) structure shows formation of additional H-bonds in the vicinity of the Ser 1110 residue, which include H-bonds between SER1110:O – :LEU1113:N, ASP1106:O – :SER1110:N, ASP1106:O – :SER1110:OG and ASP1022:OD1 – :TYR1053:OH besides prominent alterations in H-bond length affecting orientation of SER1110 in (p.Cys1110Ser) variant compared to CYS1110 in the Wt VEGFR-1 (Supplementary Figure S6). This altered H-bond pattern not only induced flexibility in the (p.Cys1110Ser) structure but also caused rearrangements of local residues with respect to solvent accessibility (Supplementary Figure S7). While TYR 1130 gets exposed to solvent, TYR 911 and ASP 1022 become buried inside. We computed the RMSF of protein backbone in order understand the structural variation in various functional motifs of VEGFR-1 owing to the (p.Cys1110Ser) variation in the distal kinase domain or activation segment of VEGFR-1. As observed from Figure 1b, the RMSF fluctuations in the variant structure were higher compared to that in the wild-type structure. The intracellular juxta membrane domain starting from residue 782–826, believed to be negative regulator of receptor activation,31 remains more or less stable and does not manifest into any structural variation in the context of distant mutational effect on conformational flexibility of VEGFR-1 intracellular domain. Our analysis based on the observed RMSF map of 860–890 and 1020–1060 stretches suggest that the Cys1110Ser variation located in the activation loop affects neighboring as well as distant residues (Figure 1b). Higher fluctuation observed across the distant residues, 860–890 stretch, contains the two important residues, namely, K of KDD at 861 and αC-Glutamate at 878 position.32, 33 Both the residues are associated with the catalytic properties of tyrosine kinase domain present in VEGFR-1 while the neighboring stretch of residues (1020–1060) that showed fluctuation fall into the catalytic loop (1020–1027) and activation segment harboring two important tyrosines, namely, TYR1048 and TYR1053, and Asparagine located at 1050, essentially serve as negative determinants that partially inhibit autophosphorylation of activation domain.34
Our findings collectively suggest that the variant structure in its expanded or relaxed form gives an easy passage for binding of ATP and other adaptor proteins and therefore the rate-determining events such as autophosphorylation and receptor activation might be occurring at higher KD values. From the signal stimulation point of view, first the protein kinases in general and VEGFR-1 in particular should undergo a conformational change on binding to the specific ligand at the cell surface, for instance, VEGF or PIGF. This conformational change is highly gauged through the αC-helix largely considered to be the part of N-terminal lobe. Our data show that the variant structure in its relaxed form attains an open conformation, which is favorable for enzyme activation and interaction with binding partners such as CBL, CRK, GRB2, SOS and so on. The glycine-rich loop (834–839 a.a, dark green) moves laterally relatively closer to the catalytic loop (1020–1027 a.a, dark yellow) in the variant structure as compared to normal structure (Figure 5a). However, the distance between the glycine-rich ATP-binding loop and the Lys 861 residue, which is responsible for forming an ion pair with alpha and beta-phosphates of ATP, remains more or less spanned at same distance (Figure 5b). This indicates that the angular cross-section of the ATP-binding cleft in wild-type and variant protein is not affected by Cys1110Ser variation and therefore the ATP passage between the glycine-rich loop and αC-helix would still be unhindered. Our study also suggests that the conformation attained by the variant structure makes it permissible for this receptor tyrosine kinase to get autophosphorylated at two sites, namely, Tyr 1048 and Tyr 1053. From the Figure 5c, it could be realized that the open conformation of the variant structure positions its activation segment in such a way that the structural steric hindrance for the phosphorylation event decreases. This alteration could also be supported by assuming that the binding pocket for substrate protein orients itself in a relaxed form, which is evident from the expanded nature of variant protein; depicted through the variation in distance between N-terminal lobe (Lys 861) and C-terminal lobe (Tyr 1048, Tyr 1053) of the variant structure, which is 27.140 and 28.096 Å as compared to wild-type structure having a constricted distance of 15.425 and 18.926 Å, respectively.
Comparison of distance spanning through the motifs of wild-type and variant tyrosine kinase domain of VEGFR-1. (a) The distance between Glycine-rich loop (green) and catalytic loop (yellow) was shortened owing to the closure of glycine loop in the variant structure; however, (b) there was no change in distance between glycine-rich loop (green) and αC-helix (brown). (c) Widening of catalytic pocket wherein the distance between the activation segment (orange) and αC-helix (brown) is significantly increased to 27.140 and 28.096 Å from the positioned Lys 861 (αC-helix) to Tyr1048 and Tyr1053 (activation loop), respectively. A full color version of this figure is available at the European Journal of Human Genetics journal online.
Having two main events modulated by Cys1110Ser variation that help in activation of VEGFR-1, depicted by atomistic MD simulation, we went on probing the effect of this conformational variability in structure on the ATP–ADP transition kinetics. Our data show that, despite binding to the same catalytic pocket, an apparently different ligand interaction pattern is observed in Wt VEGFR-1 and (p.Cys1110Ser) variant upon ATP/ADP ligand docking (Supplementary Figure S8). (p.Cys1110Ser) depicted relatively higher ATP/ADP-binding affinity coefficient compared either to that of Wt VEGFR-1 or VEGFR-2 (Figure 4, Supplementary Figure S8). Based on the ΔG score of ATP to ADP across Wt VEGFR-1, (p.Cys1110Ser) variant and VEGFR-2, it can be presumed the active kinase population (phosphorylated) in case (p.Cys1110Ser) variant is not in equilibrium state with that of inactive kinase population (unphosphorylated). This sensitivity to equilibrium shift in kinase activity could itself account for the hyperactivity of VEGFR-1 signaling in a more conventional way or by transactivation of VEGFR-2. It thus could be argued that (p.Cys1110Ser) leads to oncogenic activation of VEGFR-1 by increasing its affinity toward receptor phosphorylation.
In light of the evolved speculations about the role of VEGFR-1 in oncogenic transformations, it can be said that the variant structure attains a conformation which reinforces the transactivation of VEGFR-2 by nonspecific activation of VEGFR-1 via phosphotryosine 1048 and 1053. This crosstalk between the VEGFR-1 and VEGFR-2 could be held responsible for promoting the continual VEGFR-2-induced proliferative signal via PI3K and other players. It is also possible that the variant VEGFR-1 when excited by PLGF attenuates the negative regulation of VEGFR-2 signal module as has already been reported9 and therefore results in an increased signal output in the form of angiogenesis and cellular proliferation.
References
Shibuya M : Role of Vegf-Flt receptor system in normal and tumor angiogenesis. Adv Cancer Res 1995; 281–316.
Dvorak HF : Vascular permeability factor/vascular endothelial growth factor: a critical cytokine in tumor angiogenesis and a potential target for diagnosis and therapy. J Clin Oncol 2002; 20: 4368–4380.
Li Y : Active immunization against the vascular endothelial growth factor receptor flk1 inhibits tumor angiogenesis and metastasis. J Exp Med 2002; 195: 1575–1584.
Yoshimura K, Minami T, Nozawa M, Uemura H : Phase I clinical trial of human vascular endothelial growth factor receptor 1 peptide vaccines for patients with metastatic renal cell carcinoma. Br J Cancer 2013; 108: 1260–1266.
Shen L, Zhou Q, Wang Y et al: Antiangiogenic and antitumoral effects mediated by a vascular endothelial growth factor receptor 1 (VEGFR-1)-targeted DNAzyme. Mol Med 2013; 19: 377–386.
Olsson AK, Dimberg A, Kreuger J, Claesson-Welsh L : VEGF receptor signalling — in control of vascular function. Nat Rev Mol Cell Biol 2006; 7: 359–371.
Luttun A, Autiero M, Tjwa M, Carmeliet P : Genetic dissection of tumor angiogenesis: are PlGF and VEGFR-1 novel anti-cancer targets? Biochim Biophys Acta 2004; 1654: 79–94.
Luttun A, Tjwa M, Moons L et al: Revascularization of ischemic tissues by PlGF treatment, and inhibition of tumor angiogenesis, arthritis and atherosclerosis by anti-Flt1. Nat Med 2002; 8: 831–840.
Autiero M, Waltenberger J, Communi D et al: Role of PlGF in the intra- and intermolecular cross talk between the VEGF receptors Flt1 and Flk1. Nat Med 2003; 9: 936–943.
Carmeliet P, Moons L, Luttun A et al: Synergism between vascular endothelial growth factor and placental growth factor contributes to angiogenesis and plasma extravasation in pathological conditions. Nat Med 2001; 7: 575–583.
Ito N, Wernstedt C, Engstrom U, Claesson-Welsh L : Identification of vascular endothelial growth factor receptor-1 tyrosine phosphorylation sites and binding of SH2 domain-containing molecules. J Biol Chem 1998; 273: 23410–23418.
Sawano A, Takahashi T, Yamaguchi S, Shibuya M : The phosphorylated 1169-tyrosine containing region of Flt-1 kinase (VEGFR-1) is a major binding site for PLCγ. Biochem Biophys Res Commun 1997; 238: 487–491.
Igarashi K, Isohara T, Kato T, Shigeta K, Yamano T, Uno I : Tyrosine 1213 of Flt-1 is a major binding site of Nck and SHP-2. Biochem Biophy Res Commun 1998; 246: 95–99.
Yu Y, Hulmes JD, Herley MT, Whitney RG, Crabb JW, Sato JD : Direct identification of a major autophosphorylation site on vascular endothelial growth factor receptor Flt-1 that mediates phosphatidylinositol 3′-kinase binding. Biochem J 2001; 358: 465.
Choi Y, Sims GE, Murphy S, Miller JR, Chan AP : Predicting the functional effect of amino acid substitutions and indels. PLoS One 2012; 7: e46688.
Choi Y : A Fast Computation of Pairwise Sequence Alignment Scores Between a Protein and a Set of Single-Locus Variants of Another Protein. Proceedings of the ACM Conference on Bioinformatics, Computational Biology and Biomedicine (BCB ’12), 08–10 October 2012 ACM: New York, NY, USA, 2012, pp 414–417.
Kumar P, Henikoff S, Ng PC : Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009; 4: 1073–1081.
Adzhubei IA, Schmidt S, Peshkin L et al: A method and server for predicting damaging missense mutations. Nat Methods 2010; 7: 248–249.
Schwarz JM, Cooper DN, Schuelke M, Seelow D : MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 2014; 11: 361–362.
Accelrys Software Inc. Discovery Studio Modeling Environment, Release 3.5. San Diego, CA, USA: Accelrys Software Inc., 2012.
Hess B, Kutzner C, van der Spoel D, Lindahl E : GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 2008; 4: 435–447.
Bukhari S, Mokhdomi TA, Chikan NA et al: Affinity proteomics led identification of vimentin as a potential biomarker in colon cancers: insights from serological screening and computational modelling. Mol Biosyst 2015; 11: 159–169.
Morris GM, Huey R, Lindstrom W et al: AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 2009; 30: 2785–2791.
Vnencak-Jones C, Berger M, Pao W. Types of Molecular Tumor Testing. MyCancerGenome, 2015. Available at https://www.mycancergenome.org/content/molecular-medicine/types-of-molecular-tumor-testing/. Accessed 18 June 2015.
Ma Q, Lu AYH : Pharmacogenetics, pharmacogenomics, and individualized medicine. Pharmacol Rev 2011; 63: 437–459.
Kandoth C, McLellan MD, Vandin F et al: Mutational landscape and significance across 12 major cancer types. Nature 2013; 502: 333–339.
Cao Y : Positive and negative modulation of angiogenesis by VEGFR1 ligands. Sci Signal 2009; 2: re1.
Mimori K, Fukagawa T, Kosaka Y et al: Hematogenous metastasis in gastric cancer requires isolated tumor cells and expression of vascular endothelial growth factor receptor-1. Clin Can Res 2008; 14: 2609–2616.
Kosaka Y, Mimori K, Fukagawa T et al: Identification of the high-risk group for metastasis of gastric cancer cases by vascular endothelial growth factor receptor-1 overexpression in peripheral blood. Br J Cancer 2007; 96: 1723–1728.
Slattery ML, Lundgreen A, Wolff RK : VEGFA, FLT1, KDR and colorectal cancer: assessment of disease risk, tumor molecular phenotype, and survival. Mol Carcinog 2013; 53: e140–e150.
Gille H, Kowalski J, Yu L et al: A repressor sequence in the juxtamembrane domain of Flt-1 (VEGFR-1) constitutively inhibits vascular endothelial growth factor-dependent phosphatidylinositol 3′-kinase activation and endothelial cell migration. EMBO J 2000; 19: 4064–4073.
Fantl WJ, Johnson DE, Williams LT : Signalling by receptor tyrosine kinases. Annu Rev Biochem 1993; 62: 453–481.
Hanks SK, Quinn AM, Hunter T : The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 1988; 241: 42–52.
Meyer RD, Mohammadi M, Rahimi N : A single amino acid substitution in the activation loop defines the decoy characteristic of VEGFR-1/FLT-1. J Biol Chem 2005; 281: 867–875.
Acknowledgements
We thank Dr Omar Masood, MD, Sher-i-Kashmir Institute of Medical Sciences, Srinagar for Clinicopathological reporting; Dr Sumira Tyub, Centre of Research for Development, Srinagar for genomic data re-analysis and Dr Sarjana Dutt, Molecular Biology Division, Oncquest Laboratories Ltd for re-validation of sequencing data.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Mokhdomi, T., Bukhari, S., Chikan, N. et al. A novel kinase mutation in VEGFR-1 predisposes its αC-helix/activation loop towards allosteric activation: Atomic insights from protein simulation. Eur J Hum Genet 24, 1287–1293 (2016). https://doi.org/10.1038/ejhg.2016.26
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ejhg.2016.26
This article is cited by
-
Aqueous solution interactions with sex hormone-binding globulin and estradiol: a theoretical investigation
Journal of Biological Physics (2018)